UV-Protection Timer Controls Linkage between Stress and Pigmentation Skin Protection Systems
- PMID: 30401431
- PMCID: PMC6224604
- DOI: 10.1016/j.molcel.2018.09.022
UV-Protection Timer Controls Linkage between Stress and Pigmentation Skin Protection Systems
Abstract
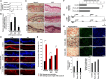
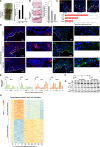
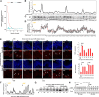
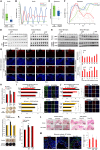
Skin sun exposure induces two protection programs: stress responses and pigmentation, the former within minutes and the latter only hours afterward. Although serving the same physiological purpose, it is not known whether and how these programs are coordinated. Here, we report that UVB exposure every other day induces significantly more skin pigmentation than the higher frequency of daily exposure, without an associated increase in stress responses. Using mathematical modeling and empirical studies, we show that the melanocyte master regulator, MITF, serves to synchronize stress responses and pigmentation and, furthermore, functions as a UV-protection timer via damped oscillatory dynamics, thereby conferring a trade-off between the two programs. MITF oscillations are controlled by multiple negative regulatory loops, one at the transcriptional level involving HIF1α and another post-transcriptional loop involving microRNA-148a. These findings support trait linkage between the two skin protection programs, which, we speculate, arose during furless skin evolution to minimize skin damage.
Keywords: MITF dynamics; UVB radiation; skin pigmentation; skin proliferation; trait linkage.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
MITF and UV responses in skin: From pigmentation to addiction.Pigment Cell Melanoma Res. 2019 Mar;32(2):224-236. doi: 10.1111/pcmr.12726. Epub 2018 Aug 3. Pigment Cell Melanoma Res. 2019. PMID: 30019545 Free PMC article. Review.
-
UVB-induced TRPS1 regulates MITF transcription activity to promote skin pigmentation.Biochim Biophys Acta Mol Basis Dis. 2024 Oct;1870(7):167445. doi: 10.1016/j.bbadis.2024.167445. Epub 2024 Jul 27. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 39074626
-
NNT mediates redox-dependent pigmentation via a UVB- and MITF-independent mechanism.Cell. 2021 Aug 5;184(16):4268-4283.e20. doi: 10.1016/j.cell.2021.06.022. Epub 2021 Jul 6. Cell. 2021. PMID: 34233163 Free PMC article.
-
A new model to investigate UVB-induced cellular senescence and pigmentation in melanocytes.Mech Ageing Dev. 2020 Sep;190:111322. doi: 10.1016/j.mad.2020.111322. Epub 2020 Jul 29. Mech Ageing Dev. 2020. PMID: 32735894 Free PMC article.
-
Skin pigmentation and its control: From ultraviolet radiation to stem cells.Exp Dermatol. 2021 Apr;30(4):560-571. doi: 10.1111/exd.14260. Epub 2020 Dec 24. Exp Dermatol. 2021. PMID: 33320376 Free PMC article. Review.
Cited by
-
Tuning Transcription Factor Availability through Acetylation-Mediated Genomic Redistribution.Mol Cell. 2020 Aug 6;79(3):472-487.e10. doi: 10.1016/j.molcel.2020.05.025. Epub 2020 Jun 11. Mol Cell. 2020. PMID: 32531202 Free PMC article.
-
MITF and TFEB cross-regulation in melanoma cells.PLoS One. 2020 Sep 3;15(9):e0238546. doi: 10.1371/journal.pone.0238546. eCollection 2020. PLoS One. 2020. PMID: 32881934 Free PMC article.
-
Construction and validation of a prognostic model for hepatocellular carcinoma: Inflammatory ferroptosis and mitochondrial metabolism indicate a poor prognosis.Front Oncol. 2023 Jan 5;12:972434. doi: 10.3389/fonc.2022.972434. eCollection 2022. Front Oncol. 2023. PMID: 36686830 Free PMC article.
-
Food-seeking behavior is triggered by skin ultraviolet exposure in males.Nat Metab. 2022 Jul;4(7):883-900. doi: 10.1038/s42255-022-00587-9. Epub 2022 Jul 11. Nat Metab. 2022. PMID: 35817855 Free PMC article.
-
New panel of biomarkers to discriminate between amelanotic and melanotic metastatic melanoma.Front Oncol. 2023 Jan 26;12:1061832. doi: 10.3389/fonc.2022.1061832. eCollection 2022. Front Oncol. 2023. PMID: 36776379 Free PMC article.
References
-
- Bayerl C., Taake S., Moll I., Jung E.G. Characterization of sunburn cells after exposure to ultraviolet light. Photodermatol. Photoimmunol. Photomed. 1995;11:149–154. - PubMed
-
- Bell R.E., Levy C. The three M’s: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 2011;24:1088–1106. - PubMed
-
- Bell R.E., Khaled M., Netanely D., Schubert S., Golan T., Buxbaum A., Janas M.M., Postolsky B., Goldberg M.S., Shamir R. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Invest. Dermatol. 2014;134:441–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

