Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: an integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging
- PMID: 30401811
- PMCID: PMC6344377
- DOI: 10.1038/s41380-018-0292-1
Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: an integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging
Abstract
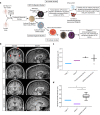
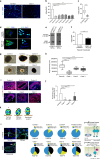
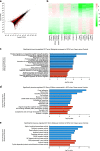
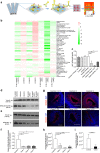
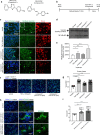
The molecular basis of how chromosome 16p13.11 microduplication leads to major psychiatric disorders is unknown. Here we have undertaken brain imaging of patients carrying microduplications in chromosome 16p13.11 and unaffected family controls, in parallel with iPS cell-derived cerebral organoid studies of the same patients. Patient MRI revealed reduced cortical volume, and corresponding iPSC studies showed neural precursor cell (NPC) proliferation abnormalities and reduced organoid size, with the NPCs therein displaying altered planes of cell division. Transcriptomic analyses of NPCs uncovered a deficit in the NFκB p65 pathway, confirmed by proteomics. Moreover, both pharmacological and genetic correction of this deficit rescued the proliferation abnormality. Thus, chromosome 16p13.11 microduplication disturbs the normal programme of NPC proliferation to reduce cortical thickness due to a correctable deficit in the NFκB signalling pathway. This is the first study demonstrating a biologically relevant, potentially ameliorable, signalling pathway underlying chromosome 16p13.11 microduplication syndrome in patient-derived neuronal precursor cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Maternally inherited autosomal dominant intellectual disability caused by 16p13.3 microduplication.Eur J Med Genet. 2016 Apr;59(4):210-4. doi: 10.1016/j.ejmg.2016.02.005. Epub 2016 Feb 9. Eur J Med Genet. 2016. PMID: 26873618
-
Concurrent de novo MACF1 mutation and inherited 16p13.11 microduplication in a preterm newborn with hypotonia, joint hyperlaxity and multiple congenital malformations: a case report.BMC Pediatr. 2024 Aug 16;24(1):528. doi: 10.1186/s12887-024-04628-y. BMC Pediatr. 2024. PMID: 39152427 Free PMC article.
-
A microduplication of the Rubinstein-Taybi region on 16p13.3 in a girl with a bilateral complete cleft lip and palate and severe mental retardation.Clin Dysmorphol. 2012 Oct;21(4):204-207. doi: 10.1097/MCD.0b013e3283554d15. Clin Dysmorphol. 2012. PMID: 22664659 No abstract available.
-
16p13.11 microduplication with growth retardation and developmental disorders: a case report and literature review.Nagoya J Med Sci. 2025 Feb;87(1):144-149. doi: 10.18999/nagjms.87.1.144. Nagoya J Med Sci. 2025. PMID: 40256007 Free PMC article. Review.
-
Interstitial 16p13.3 microduplication: case report and critical review of genotype-phenotype correlation.Eur J Med Genet. 2012 Dec;55(12):747-52. doi: 10.1016/j.ejmg.2012.09.006. Epub 2012 Sep 29. Eur J Med Genet. 2012. PMID: 23032921 Review.
Cited by
-
The Multifaceted Roles of Primary Cilia in the Development of the Cerebral Cortex.Front Cell Dev Biol. 2021 Feb 2;9:630161. doi: 10.3389/fcell.2021.630161. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33604340 Free PMC article. Review.
-
Altered neuroepithelial morphogenesis and migration defects in iPSC-derived cerebral organoids and 2D neural stem cells in familial bipolar disorder.Oxf Open Neurosci. 2024 Apr 3;3:kvae007. doi: 10.1093/oons/kvae007. eCollection 2024. Oxf Open Neurosci. 2024. PMID: 38638145 Free PMC article.
-
Cellular Models in Schizophrenia Research.Int J Mol Sci. 2021 Aug 7;22(16):8518. doi: 10.3390/ijms22168518. Int J Mol Sci. 2021. PMID: 34445221 Free PMC article. Review.
-
What neuroscience has already done for us.BJPsych Bull. 2020 Jun;44(3):110-112. doi: 10.1192/bjb.2019.90. BJPsych Bull. 2020. PMID: 32077401 Free PMC article.
-
Building the brain from scratch: Engineering region-specific brain organoids from human stem cells to study neural development and disease.Curr Top Dev Biol. 2021;142:477-530. doi: 10.1016/bs.ctdb.2020.12.011. Epub 2021 Feb 18. Curr Top Dev Biol. 2021. PMID: 33706925 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- SCD/12/CSO_/Chief Scientist Office/United Kingdom
- NC/C011202/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- MR/J004367/1/MRC_/Medical Research Council/United Kingdom
- 100135/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

