Onsite GTP fuelling via DYNAMO1 drives division of mitochondria and peroxisomes
- PMID: 30401830
- PMCID: PMC6219506
- DOI: 10.1038/s41467-018-07009-z
Onsite GTP fuelling via DYNAMO1 drives division of mitochondria and peroxisomes
Abstract
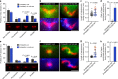
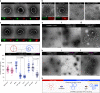
Mitochondria and peroxisomes proliferate by division. During division, a part of their membrane is pinched off by constriction of the ring-shaped mitochondrial division (MD) and peroxisome-dividing (POD) machinery. This constriction is mediated by a dynamin-like GTPase Dnm1 that requires a large amount of GTP as an energy source. Here, via proteomics of the isolated division machinery, we show that the 17-kDa nucleoside diphosphate kinase-like protein, dynamin-based ring motive-force organizer 1 (DYNAMO1), locally generates GTP in MD and POD machineries. DYNAMO1 is widely conserved among eukaryotes and colocalizes with Dnm1 on the division machineries. DYNAMO1 converts ATP to GTP, and disruption of its activity impairs mitochondrial and peroxisomal fissions. DYNAMO1 forms a ring-shaped complex with Dnm1 and increases the magnitude of the constricting force. Our results identify DYNAMO1 as an essential component of MD and POD machineries, suggesting that local GTP generation in Dnm1-based machinery regulates motive force for membrane severance.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

