Directed Supramolecular Organization of N-BAR Proteins through Regulation of H0 Membrane Immersion Depth
- PMID: 30401832
- PMCID: PMC6219572
- DOI: 10.1038/s41598-018-34273-2
Directed Supramolecular Organization of N-BAR Proteins through Regulation of H0 Membrane Immersion Depth
Abstract
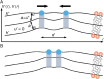
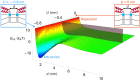
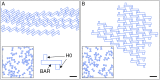
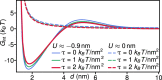
Many membrane remodeling events rely on the ability of curvature-generating N-BAR membrane proteins to organize into distinctive supramolecular configurations. Experiments have revealed a conformational switch in N-BAR proteins resulting in vesicular or tubular membrane shapes, with shallow membrane immersion of the H0 amphipathic helices of N-BAR proteins on vesicles but deep H0 immersion on tubes. We develop here a minimal elastic model of the local thinning of the lipid bilayer resulting from H0 immersion. Our model predicts that the observed conformational switch in N-BAR proteins produces a corresponding switch in the bilayer-mediated N-BAR interactions due to the H0 helices. In agreement with experiments, we find that bilayer-mediated H0 interactions oppose N-BAR multimerization for the shallow H0 membrane immersion depths measured on vesicles, but promote self-assembly of supramolecular N-BAR chains for the increased H0 membrane immersion depths measured on tubes. Finally, we consider the possibility that bilayer-mediated H0 interactions might contribute to the concerted structural reorganization of N-BAR proteins suggested by experiments. Our results indicate that the membrane immersion depth of amphipathic protein helices may provide a general molecular control parameter for membrane organization.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Role of helix 0 of the N-BAR domain in membrane curvature generation.Biophys J. 2008 Apr 15;94(8):3065-73. doi: 10.1529/biophysj.107.113118. Epub 2008 Jan 16. Biophys J. 2008. PMID: 18199667 Free PMC article.
-
Understanding the role of amphipathic helices in N-BAR domain driven membrane remodeling.Biophys J. 2013 Jan 22;104(2):404-11. doi: 10.1016/j.bpj.2012.12.006. Biophys J. 2013. PMID: 23442862 Free PMC article.
-
Factors influencing local membrane curvature induction by N-BAR domains as revealed by molecular dynamics simulations.Biophys J. 2008 Aug;95(4):1866-76. doi: 10.1529/biophysj.107.121160. Epub 2008 May 9. Biophys J. 2008. PMID: 18469070 Free PMC article.
-
BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature.Biochim Biophys Acta. 2006 Aug;1761(8):897-912. doi: 10.1016/j.bbalip.2006.06.015. Epub 2006 Jul 28. Biochim Biophys Acta. 2006. PMID: 16938488 Review.
-
A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins.Semin Cell Dev Biol. 2010 Jun;21(4):381-90. doi: 10.1016/j.semcdb.2009.12.004. Epub 2009 Dec 16. Semin Cell Dev Biol. 2010. PMID: 20006726 Review.
Cited by
-
Generation of nanoscopic membrane curvature for membrane trafficking.Nat Rev Mol Cell Biol. 2023 Jan;24(1):63-78. doi: 10.1038/s41580-022-00511-9. Epub 2022 Aug 2. Nat Rev Mol Cell Biol. 2023. PMID: 35918535 Review.
-
Acute diacylglycerol production activates critical membrane-shaping proteins leading to mitochondrial tubulation and fission.Nat Commun. 2025 Mar 19;16(1):2685. doi: 10.1038/s41467-025-57439-9. Nat Commun. 2025. PMID: 40102394 Free PMC article.
-
Membrane Proteins and Membrane Curvature: Mutual Interactions and a Perspective on Disease Treatments.Biomolecules. 2023 Dec 11;13(12):1772. doi: 10.3390/biom13121772. Biomolecules. 2023. PMID: 38136643 Free PMC article. Review.
-
Annexin B12 Trimer Formation is Governed by a Network of Protein-Protein and Protein-Lipid Interactions.Sci Rep. 2020 Mar 24;10(1):5301. doi: 10.1038/s41598-020-62343-x. Sci Rep. 2020. PMID: 32210350 Free PMC article.
-
Membrane Remodeling Driven by Shallow Helix Insertions via a Cooperative Mechanism.Membranes (Basel). 2025 Apr 1;15(4):101. doi: 10.3390/membranes15040101. Membranes (Basel). 2025. PMID: 40277971 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

