High-level expression and molecular characterization of a recombinant prolidase from Escherichia coli NovaBlue
- PMID: 30402354
- PMCID: PMC6215446
- DOI: 10.7717/peerj.5863
High-level expression and molecular characterization of a recombinant prolidase from Escherichia coli NovaBlue
Abstract
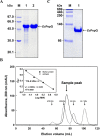
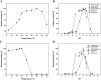
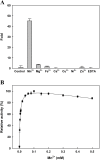
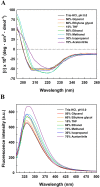
Long-term use of organophosphorus (OP) compounds has become an increasing global problem and a major threat to sustainability and human health. Prolidase is a proline-specific metallopeptidase that can offer an efficient option for the degradation of OP compounds. In this study, a full-length gene from Escherichia coli NovaBlue encoding a prolidase (EcPepQ) was amplified and cloned into the commercially-available vector pQE-30 to yield pQE-EcPepQ. The overexpressed enzyme was purified from the cell-free extract of isopropyl thio-β-D-galactoside IPTG-induced E. coli M15 (pQE-EcPepQ) cells by nickel-chelate chromatography. The molecular mass of EcPepQ was determined to be about 57 kDa by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the result of size-exclusion chromatography demonstrated that the enzyme was mainly present in 25 mM Tris-HCl buffer (pH 8.0) as a dimeric form. The optimal conditions for EcPepQ activity were 60 °C, pH 8.0, and 0.1 mM Mn2+ ion. Kinetic analysis with Ala-Pro as the substrate showed that the K m and k cat values of EcPepQ were 8.8 mM and 926.5 ± 2.0 s-1, respectively. The thermal unfolding of EcPepQ followed a two-state process with one well-defined unfolding transition of 64.2 °C. Analysis of guanidine hydrochloride (GdnHCl)-induced denaturation by tryptophan emission fluorescence spectroscopy revealed that the enzyme had a [GdnHCl]0.5,N-U value of 1.98 M. The purified enzyme also exhibited some degree of tolerance to various water/organic co-solvents. Isopropanol and tetrahydrofuran were very detrimental to the enzymatic activity of EcPepQ; however, other more hydrophilic co-solvents, such as formamide, methanol, and ethylene glycol, were better tolerated. Eventually, the non-negative influence of some co-solvents on both catalytic activity and structural stability of EcPepQ allows to adjust the reaction conditions more suitable for EcPepQ-catalyzed bioprocess.
Keywords: Chaotropic agent-induced denaturation; Escherichia coli; Gene expression; Molecular characterization; Organic co-solvents; Prolidase.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Affinity Immobilization of a Bacterial Prolidase onto Metal-Ion-Chelated Magnetic Nanoparticles for the Hydrolysis of Organophosphorus Compounds.Int J Mol Sci. 2019 Jul 24;20(15):3625. doi: 10.3390/ijms20153625. Int J Mol Sci. 2019. PMID: 31344929 Free PMC article.
-
Expression optimization and biochemical characterization of a recombinant gamma-glutamyltranspeptidase from Escherichia coli novablue.Protein J. 2006 Sep;25(6):431-41. doi: 10.1007/s10930-006-9037-0. Protein J. 2006. PMID: 17094029
-
Overexpression of a recombinant gamma-glutamyltranspeptidase from Escherichia coli Novablue.Indian J Biochem Biophys. 2006 Dec;43(6):345-50. Indian J Biochem Biophys. 2006. PMID: 17285798
-
Overexpression, one-step purification, and biochemical characterization of a recombinant gamma-glutamyltranspeptidase from Bacillus licheniformis.Appl Microbiol Biotechnol. 2006 Nov;73(1):103-12. doi: 10.1007/s00253-006-0440-4. Epub 2006 Jun 21. Appl Microbiol Biotechnol. 2006. PMID: 16850301
-
Molecular characterization of a novel trehalose-6-phosphate hydrolase, TreA, from Bacillus licheniformis.Int J Biol Macromol. 2012 Apr 1;50(3):459-70. doi: 10.1016/j.ijbiomac.2012.01.011. Epub 2012 Jan 18. Int J Biol Macromol. 2012. PMID: 22285990
Cited by
-
Affinity Immobilization of a Bacterial Prolidase onto Metal-Ion-Chelated Magnetic Nanoparticles for the Hydrolysis of Organophosphorus Compounds.Int J Mol Sci. 2019 Jul 24;20(15):3625. doi: 10.3390/ijms20153625. Int J Mol Sci. 2019. PMID: 31344929 Free PMC article.
References
-
- Browne P, O’Cuinn G. The purification and characterization of a proline dipeptidase from guinea pig brain. Journal of Biological Chemistry. 1983;258:6147–6154. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

