In silico approach to identify non-synonymous SNPs with highest predicted deleterious effect on protein function in human obesity related gene, neuronal growth regulator 1 (NEGR1)
- PMID: 30402368
- PMCID: PMC6212372
- DOI: 10.1007/s13205-018-1463-0
In silico approach to identify non-synonymous SNPs with highest predicted deleterious effect on protein function in human obesity related gene, neuronal growth regulator 1 (NEGR1)
Abstract
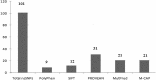
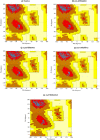
Neuronal growth regulator 1 (NEGR1) is a candidate gene for human obesity, which encodes the neural cell adhesion and growth molecule. The aim of the current study was to recognize the non-synonymous SNPs (nsSNPs) with the highest predicted deleterious effect on protein function of the NEGR1 gene. We have used five computational tools, namely, PolyPhen, SIFT, PROVEAN, MutPred and M-CAP, to predict the deleterious and pathogenic nsSNPs of the NEGR1 gene. Homology modeling approach was used to model the native and mutant NEGR1 protein models. Furthermore, structural validation was performed by the PROCHECK server to interpret the stability of the predicted models. We have predicted four potential deleterious nsSNPs, i.e., rs145524630 (Ala70Thr), rs267598710 (Pro168Leu), rs373419972 (Arg239Cys) and rs375352213 (Leu158Phe), which might be involved in causing obesity phenotypes. The predicted mutant models showed higher root mean square deviation and free energy values under the PyMoL and SWISS-PDB viewer, respectively. Additionally, the FTSite server predicted one nsSNP, i.e., rs145524630 (Ala70Thr) out of four identified nsSNPs found in the NEGR1 protein-binding site. There were four potential deleterious and pathogenic nsSNPs, i.e., rs145524630, rs267598710, rs373419972 and rs375352213, identified from the above-mentioned tools. In future, further functional in vitro and in vivo analysis could lead to better knowledge about these nsSNPs on the influence of the NEGR1 gene in causing human obesity. Hence, the present computational examination suggest that predicated nsSNPs may feasibly be a drug target and play an important role in contributing to human obesity.
Keywords: Computational tools; Homology modeling; NEGR1; Obesity; nsSNPs.
Conflict of interest statement
Compliance with ethical standardsThe authors declare no conflicts of interest for publishing this work.
Figures

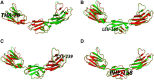

Similar articles
-
In silico approach to identify non-synonymous SNPs in human obesity related gene, MC3R (melanocortin-3-receptor).Comput Biol Chem. 2017 Apr;67:122-130. doi: 10.1016/j.compbiolchem.2016.12.009. Epub 2016 Dec 29. Comput Biol Chem. 2017. PMID: 28073065
-
Determination of deleterious single-nucleotide polymorphisms of human LYZ C gene: an in silico study.J Genet Eng Biotechnol. 2022 Jul 1;20(1):92. doi: 10.1186/s43141-022-00383-8. J Genet Eng Biotechnol. 2022. PMID: 35776277 Free PMC article.
-
Predicting the functional and structural consequences of nsSNPs in human methionine synthase gene using computational tools.Syst Biol Reprod Med. 2019 Aug;65(4):288-300. doi: 10.1080/19396368.2019.1568611. Epub 2019 Jan 24. Syst Biol Reprod Med. 2019. PMID: 30676783
-
Screening and insilico analysis of deleterious nsSNPs (missense) in human CSF3 for their effects on protein structure, stability and function.Comput Biol Chem. 2019 Oct;82:57-64. doi: 10.1016/j.compbiolchem.2019.06.001. Epub 2019 Jun 6. Comput Biol Chem. 2019. PMID: 31272062
-
From Bound Cells Comes a Sound Mind: The Role of Neuronal Growth Regulator 1 in Psychiatric Disorders.Exp Neurobiol. 2020 Feb 29;29(1):1-10. doi: 10.5607/en.2020.29.1.1. Exp Neurobiol. 2020. PMID: 32122104 Free PMC article. Review.
Cited by
-
In silico profiling of nonsynonymous SNPs of fat mass and obesity-associated gene: possible impacts on the treatment of non-alcoholic fatty liver disease.Lipids Health Dis. 2023 Jan 30;22(1):17. doi: 10.1186/s12944-023-01782-7. Lipids Health Dis. 2023. PMID: 36717943 Free PMC article.
-
Identification of High-Risk Single Nucleotide Polymorphisms in the Human CYB5R3 Gene Responsible for Recessive Congenital Methemoglobinemia: A Computational Approach.Mol Syndromol. 2023 Oct;14(5):375-393. doi: 10.1159/000530173. Epub 2023 May 5. Mol Syndromol. 2023. PMID: 37901856 Free PMC article.
-
Computational Analysis of IDH1, IDH2, and TP53 Mutations in Low-Grade Gliomas Including Oligodendrogliomas and Astrocytomas.Cancer Inform. 2020 Apr 15;19:1176935120915839. doi: 10.1177/1176935120915839. eCollection 2020. Cancer Inform. 2020. PMID: 32313423 Free PMC article.
References
-
- Alonso R, Farías M, Alvarez V, Cuevas A. Translational cardiometabolic genomic medicine. Amsterdam: Elsevier; 2016. The genetics of obesity; pp. 161–177.
LinkOut - more resources
Full Text Sources
Miscellaneous

