Onco-Multi-OMICS Approach: A New Frontier in Cancer Research
- PMID: 30402498
- PMCID: PMC6192166
- DOI: 10.1155/2018/9836256
Onco-Multi-OMICS Approach: A New Frontier in Cancer Research
Abstract
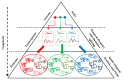
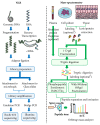
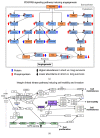
The acquisition of cancer hallmarks requires molecular alterations at multiple levels including genome, epigenome, transcriptome, proteome, and metabolome. In the past decade, numerous attempts have been made to untangle the molecular mechanisms of carcinogenesis involving single OMICS approaches such as scanning the genome for cancer-specific mutations and identifying altered epigenetic-landscapes within cancer cells or by exploring the differential expression of mRNA and protein through transcriptomics and proteomics techniques, respectively. While these single-level OMICS approaches have contributed towards the identification of cancer-specific mutations, epigenetic alterations, and molecular subtyping of tumors based on gene/protein-expression, they lack the resolving-power to establish the casual relationship between molecular signatures and the phenotypic manifestation of cancer hallmarks. In contrast, the multi-OMICS approaches involving the interrogation of the cancer cells/tissues in multiple dimensions have the potential to uncover the intricate molecular mechanism underlying different phenotypic manifestations of cancer hallmarks such as metastasis and angiogenesis. Moreover, multi-OMICS approaches can be used to dissect the cellular response to chemo- or immunotherapy as well as discover molecular candidates with diagnostic/prognostic value. In this review, we focused on the applications of different multi-OMICS approaches in the field of cancer research and discussed how these approaches are shaping the field of personalized oncomedicine. We have highlighted pioneering studies from "The Cancer Genome Atlas (TCGA)" consortium encompassing integrated OMICS analysis of over 11,000 tumors from 33 most prevalent forms of cancer. Accumulation of huge cancer-specific multi-OMICS data in repositories like TCGA provides a unique opportunity for the systems biology approach to tackle the complexity of cancer cells through the unification of experimental data and computational/mathematical models. In future, systems biology based approach is likely to predict the phenotypic changes of cancer cells upon chemo-/immunotherapy treatment. This review is sought to encourage investigators to bring these different approaches together for interrogating cancer at molecular, cellular, and systems levels.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

