A single-cell survey of the human first-trimester placenta and decidua
- PMID: 30402542
- PMCID: PMC6209386
- DOI: 10.1126/sciadv.aau4788
A single-cell survey of the human first-trimester placenta and decidua
Abstract
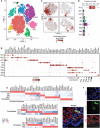
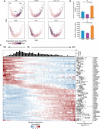
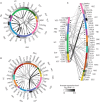
The placenta and decidua interact dynamically to enable embryonic and fetal development. Here, we report single-cell RNA sequencing of 14,341 and 6754 cells from first-trimester human placental villous and decidual tissues, respectively. Bioinformatic analysis identified major cell types, many known and some subtypes previously unknown in placental villi and decidual context. Further detailed analysis revealed proliferating subpopulations, enrichment of cell type-specific transcription factors, and putative intercellular communication in the fetomaternal microenvironment. This study provides a blueprint to further the understanding of the roles of these cells in the placenta and decidua for maintenance of early gestation as well as pathogenesis in pregnancy-related disorders.
Figures


References
-
- Maltepe E., Fisher S. J., Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523–552 (2015). - PubMed
-
- Garrido-Gomez T., Dominguez F., Quiñonero A., Diaz-Gimeno P., Kapidzic M., Gormley M., Ona K., Padilla-Iserte P., McMaster M., Genbacev O., Perales A., Fisher S. J., Simón C., Defective decidualization during and after severe preeclampsia reveals a possible maternal contribution to the etiology. Proc. Natl. Acad. Sci. U.S.A. 114, E8468–E8477 (2017). - PMC - PubMed
-
- Norwitz E. R., Schust D. J., Fisher S. J., Implantation and the survival of early pregnancy. N. Engl. J. Med. 345, 1400–1408 (2001). - PubMed
-
- Gude N. M., Roberts C. T., Kalionis B., King R. G., Growth and function of the normal human placenta. Thromb. Res. 114, 397–407 (2004). - PubMed
-
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., McCarroll S. A., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

