DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell-related genes in mouse embryonic stem cells
- PMID: 30403691
- PMCID: PMC6221296
- DOI: 10.1371/journal.pone.0205969
DNMTs and SETDB1 function as co-repressors in MAX-mediated repression of germ cell-related genes in mouse embryonic stem cells
Abstract
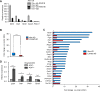
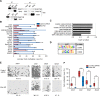
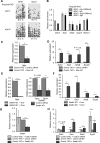
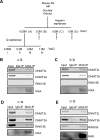
In embryonic stem cells (ESCs), the expression of development-related genes, including germ cell-related genes, is globally repressed. The transcription factor MAX represses germ cell-related gene expression in ESCs via PCGF6-polycomb repressive complex 1 (PRC1), which consists of several epigenetic factors. However, we predicted that MAX represses germ cell-related gene expression through several additional mechanisms because PCGF6-PRC1 regulates the expression of only a subset of genes repressed by MAX. Here, we report that MAX associated with DNA methyltransferases (DNMTs) and the histone methyltransferase SETDB1 cooperatively control germ cell-related gene expression in ESCs. Both DNA methylation and histone H3 lysine 9 tri-methylation of the promoter regions of several germ cell-related genes were not affected by knockout of the PRC1 components, indicating that the MAX-DNMT and MAX-SETDB1 pathways are independent of the PCGF6-PRC1 pathway. Our findings provide insights into our understanding of MAX-based repressive mechanisms of germ cell-related genes in ESCs.
Conflict of interest statement
IM is employed by and received salary from HaploPharma Inc., Chuo-ku, Tokyo, Japan. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials.
Figures

Similar articles
-
Repression of germline genes by PRC1.6 and SETDB1 in the early embryo precedes DNA methylation-mediated silencing.Nat Commun. 2021 Dec 2;12(1):7020. doi: 10.1038/s41467-021-27345-x. Nat Commun. 2021. PMID: 34857746 Free PMC article.
-
The polycomb group protein PCGF6 mediates germline gene silencing by recruiting histone-modifying proteins to target gene promoters.J Biol Chem. 2020 Jul 10;295(28):9712-9724. doi: 10.1074/jbc.RA119.012121. Epub 2020 Jun 1. J Biol Chem. 2020. PMID: 32482889 Free PMC article.
-
Max is a repressor of germ cell-related gene expression in mouse embryonic stem cells.Nat Commun. 2013;4:1754. doi: 10.1038/ncomms2780. Nat Commun. 2013. PMID: 23612295
-
The functions of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) in biological process and disease.Epigenetics Chromatin. 2023 Dec 7;16(1):47. doi: 10.1186/s13072-023-00519-1. Epigenetics Chromatin. 2023. PMID: 38057834 Free PMC article. Review.
-
Histone lysine methylation in genomic imprinting.Epigenetics. 2009 May 16;4(4):216-20. doi: 10.4161/epi.8974. Epub 2009 May 11. Epigenetics. 2009. PMID: 19483465 Review.
Cited by
-
Unreprogrammed H3K9me3 prevents minor zygotic genome activation and lineage commitment in SCNT embryos.Nat Commun. 2023 Aug 9;14(1):4807. doi: 10.1038/s41467-023-40496-3. Nat Commun. 2023. PMID: 37558707 Free PMC article.
-
On transposons and totipotency.Philos Trans R Soc Lond B Biol Sci. 2020 Mar 30;375(1795):20190339. doi: 10.1098/rstb.2019.0339. Epub 2020 Feb 10. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32075562 Free PMC article.
-
MGA directly recruits SETDB1/ATF7IP for histone H3K9me3 mark on meiosis-related genes in mouse embryonic stem cells.iScience. 2025 Jul 5;28(8):113059. doi: 10.1016/j.isci.2025.113059. eCollection 2025 Aug 15. iScience. 2025. PMID: 40727931 Free PMC article.
-
Crosstalk within and beyond the Polycomb repressive system.J Cell Biol. 2024 May 6;223(5):e202311021. doi: 10.1083/jcb.202311021. Epub 2024 Mar 20. J Cell Biol. 2024. PMID: 38506728 Free PMC article. Review.
-
Kaiso Regulates DNA Methylation Homeostasis.Int J Mol Sci. 2021 Jul 15;22(14):7587. doi: 10.3390/ijms22147587. Int J Mol Sci. 2021. PMID: 34299205 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

