Transmissibility of emerging viral zoonoses
- PMID: 30403733
- PMCID: PMC6221319
- DOI: 10.1371/journal.pone.0206926
Transmissibility of emerging viral zoonoses
Abstract
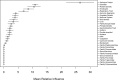
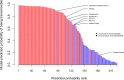
Effective public health research and preparedness requires an accurate understanding of which virus species possess or are at risk of developing human transmissibility. Unfortunately, our ability to identify these viruses is limited by gaps in disease surveillance and an incomplete understanding of the process of viral adaptation. By fitting boosted regression trees to data on 224 human viruses and their associated traits, we developed a model that predicts the human transmission ability of zoonotic viruses with over 84% accuracy. This model identifies several viruses that may have an undocumented capacity for transmission between humans. Viral traits that predicted human transmissibility included infection of nonhuman primates, the absence of a lipid envelope, and detection in the human nervous system and respiratory tract. This predictive model can be used to prioritize high-risk viruses for future research and surveillance, and could inform an integrated early warning system for emerging infectious diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Morse S. S. & Schluederberg A. Emerging Viruses: The Evolution of Viruses and Viral Diseases. J. Infect. Dis. 162, 1–7 (1990). - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

