Annotation and analysis of the mitochondrial genome of Coniothyrium glycines, causal agent of red leaf blotch of soybean, reveals an abundance of homing endonucleases
- PMID: 30403741
- PMCID: PMC6221350
- DOI: 10.1371/journal.pone.0207062
Annotation and analysis of the mitochondrial genome of Coniothyrium glycines, causal agent of red leaf blotch of soybean, reveals an abundance of homing endonucleases
Abstract
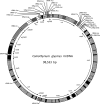
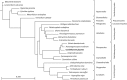
Coniothyrium glycines, the causal agent of soybean red leaf blotch, is a USDA APHIS-listed Plant Pathogen Select Agent and potential threat to US agriculture. Sequencing of the C. glycines mt genome revealed a circular 98,533-bp molecule with a mean GC content of 29.01%. It contains twelve of the mitochondrial genes typically involved in oxidative phosphorylation (atp6, cob, cox1-3, nad1-6, and nad4L), one for a ribosomal protein (rps3), four for hypothetical proteins, one for each of the small and large subunit ribosomal RNAs (rns and rnl) and a set of 30 tRNAs. Genes were encoded on both DNA strands with cox1 and cox2 occurring as adjacent genes having no intergenic spacers. Likewise, nad2 and nad3 are adjacent with no intergenic spacers and nad5 is immediately followed by nad4L with an overlap of one base. Thirty-two introns, comprising 54.1% of the total mt genome, were identified within eight protein-coding genes and the rnl. Eighteen of the introns contained putative intronic ORFs with either LAGLIDADG or GIY-YIG homing endonuclease motifs, and an additional eleven introns showed evidence of truncated or degenerate endonuclease motifs. One intron possessed a degenerate N-acetyl-transferase domain. C. glycines shares some conservation of gene order with other members of the Pleosporales, most notably nad6-rnl-atp6 and associated conserved tRNA clusters. Phylogenetic analysis of the twelve shared protein coding genes agrees with commonly accepted fungal taxonomy. C. glycines represents the second largest mt genome from a member of the Pleosporales sequenced to date. This research provides the first genomic information on C. glycines, which may provide targets for rapid diagnostic assays and population studies.
Conflict of interest statement
BC and RAW are employees of MRIGlobal (www.mriglobal.org). The commercial affiliation of BC and RAW does not alter our adherence to all PLOS ONE policies on sharing data and materials, and there are no patents, products in development or marketed products to declare. All other authors declare no competing interests.
Figures

References
-
- Hartman GL, Sinclair JB. Dactuliochaeta, a new genus for the fungus causing red leaf blotch of soybeans. Mycologia. 1988;80(5):696–706.
-
- Punithalingam E. CMI descriptions of fungi and bacteria: Dactuliochaeta glycines (No. 1012). 1990.
-
- Datnoff LE, Naik DM, Sinclair JB. Effect of red leaf blotch on soybean yields in Zambia. Plant Dis. 1987;71:132–135.
-
- Hartman GL, Sinclair JB. Red leaf blotch (Dactuliochaeta glycines) of soybeans (Glycine max) and its relationship to yield. Plant Pathol. 1996;45(2):332–343.
-
- Hartman GL, Sinclair JB. Cultural studies on Dactuliochaeta glycines, the causal agent of red leaf blotch of soybeans. Plant Dis. 1992;76(8):847–852.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

