Identification of the novel Ido1 imprinted locus and its potential epigenetic role in pregnancy loss
- PMID: 30403776
- PMCID: PMC6360327
- DOI: 10.1093/hmg/ddy383
Identification of the novel Ido1 imprinted locus and its potential epigenetic role in pregnancy loss
Abstract
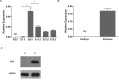
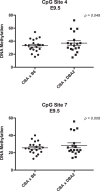
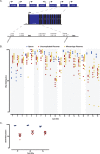
Previous studies show that aberrant tryptophan catabolism reduces maternal immune tolerance and adversely impacts pregnancy outcomes. Tryptophan depletion in pregnancy is facilitated by increased activity of tryptophan-depleting enzymes [i.e. the indolamine-2,3 dioxygenase (IDO)1 and IDO2) in the placenta. In mice, inhibition of IDO1 activity during pregnancy results in fetal loss; however, despite its important role, regulation of Ido1 gene transcription is unknown. The current study shows that the Ido1 and Ido2 genes are imprinted and maternally expressed in mouse placentas. DNA methylation analysis demonstrates that nine CpG sites at the Ido1 promoter constitute a differentially methylated region that is highly methylated in sperm but unmethylated in oocytes. Bisulfite cloning sequencing analysis shows that the paternal allele is hypermethylated while the maternal allele shows low levels of methylation in E9.5 placenta. Further study in E9.5 placentas from the CBA/J X DBA/2 spontaneous abortion mouse model reveals that aberrant methylation of Ido1 is linked to pregnancy loss. DNA methylation analysis in humans shows that IDO1 is hypermethylated in human sperm but partially methylated in placentas, suggesting similar methylation patterns to mouse. Importantly, analysis in euploid placentas from first trimester pregnancy loss reveals that IDO1 methylation significantly differs between the two placenta cohorts, with most CpG sites showing increased percent of methylation in miscarriage placentas. Our study suggests that DNA methylation is linked to regulation of Ido1/IDO1 expression and altered Ido1/IDO1 DNA methylation can adversely influence pregnancy outcomes.
Figures


References
-
- Alegre E., López A.S., Díaz-Lagares A. and González A. (2008) Study of the plasmatic levels of tryptophan and kynurenine throughout pregnancy. Clin. Chim. Acta, 393, 132–133. - PubMed
-
- Aluvihare V.R., Kallikourdis M. and Betz A.G. (2004) Regulatory T cells mediate maternal tolerance to the fetus. Nat. Immunol., 5, 266–271. - PubMed
-
- Arefayene M., Philips S., Cao D., Mamidipalli S., Desta Z., Flockhart D.A., Wilkes D.S. and Skaar T.C. (2009) Identification of genetic variants in the human indoamine 2,3-dioxygenase (IDO1) gene, which have altered enzyme activity. Pharmacogenet. Genomics, 19 (6), 464–476. - PubMed
-
- Baban B., Chandler P., McCool D., Marshall B., Munn D.H. and Mellor A.L. (2004) Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. J. Reprod. Immunol., 61, 67–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

