Recombination and Large Structural Variations Shape Interspecific Edible Bananas Genomes
- PMID: 30403808
- PMCID: PMC6340459
- DOI: 10.1093/molbev/msy199
Recombination and Large Structural Variations Shape Interspecific Edible Bananas Genomes
Abstract
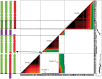
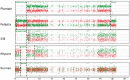
Admixture and polyploidization are major recognized eukaryotic genome evolutionary processes. Their impacts on genome dynamics vary among systems and are still partially deciphered. Many banana cultivars are triploid (sometimes diploid) interspecific hybrids between Musa acuminata (A genome) and M. balbisiana (B genome). They have no or very low fertility, are vegetatively propagated and have been classified as "AB," "AAB," or "ABB" based on morphological characters. We used NGS sequence data to characterize the A versus B chromosome composition of nine diploid and triploid interspecific cultivars, to compare the chromosome structures of A and B genomes and analyze A/B chromosome segregations in a polyploid context. We showed that interspecific recombination occurred frequently between A and B chromosomes. We identified two large structural variations between A and B genomes, a reciprocal translocation and an inversion that locally affected recombination and led to segregation distortion and aneuploidy in a triploid progeny. Interspecific recombination and large structural variations explained the mosaic genomes observed in edible bananas. The unprecedented resolution in deciphering their genome structure allowed us to start revisiting the origins of banana cultivars and provided new information to gain insight into the impact of interspecificity on genome evolution. It will also facilitate much more effective assessment of breeding strategies.
Figures




References
-
- Boonruangrod R, Desai D, Fluch S, Berenyi M, Burg K.. 2008. Identification of cytoplasmic ancestor gene-pools of Musa acuminata Colla and Musa balbisiana Colla and their hybrids by chloroplast and mitochondrial haplotyping. Theor Appl Genet. 118(1):43–55. - PubMed
-
- Carreel F, de Leon DG, Lagoda P, Lanaud C, Jenny C, Horry JP, du Montcel HT.. 2002. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45(4):679–692. - PubMed
-
- Carreel F, Fauré S, De Leon DG, Lagoda PJL, Perrier X, Bakry F, Du Montcel HT, Lanaud C, Horry J-P.. 1994. Evaluation de la diversité génétique chez les bananiers diploïdes (Musa sp.). Genet Sel Evol. 26(Suppl):125s–136s.
-
- Cheesman EE. 1947. Classification of the bananas. I. The genus Ensete Horan and the genus Musa L. Kew Bulletin 2(2):97–117.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

