Discovery of CDK8/CycC Ligands with a New Virtual Screening Tool
- PMID: 30403831
- PMCID: PMC6428209
- DOI: 10.1002/cmdc.201800559
Discovery of CDK8/CycC Ligands with a New Virtual Screening Tool
Abstract
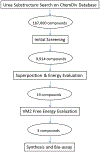
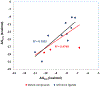
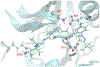
Selective inhibition of cyclin-dependent kinase 8 and cyclin C (CDK8/CycC) has been suggested as a promising strategy for decreasing mitogenic signals in cancer cells with reduced toxicity toward normal cells. We developed a novel virtual screening protocol for drug development and applied it to the discovery of new CDK8/CycC type II ligands, which is likely to achieve long residence time and specificity. We first analyzed the binding thermodynamics of 11 published pyrazolourea ligands using molecular dynamics simulations and a free-energy calculation method, VM2, and extracted the key binding information to assist virtual screening. The urea moiety was found to be the critical structural contributor of the reference ligands. Starting with the urea moiety, we conducted substructure-based searches with our newly developed superposition and single-point energy evaluation method, followed by free-energy calculations, and singled out three purchasable compounds for bioassay testing. The ranking from the experimental results is completely consistent with the predicted rankings. A potent drug-like compound was found to have a Kd value of 42.5 nm, which is similar to those of the most potent reference ligands; this provided a good starting point for further improvement. This study shows that our novel virtual screening protocol is an accurate and efficient tool for drug development.
Keywords: binding affinity; compound fragments; computer simulations; in silico screening; kinetics; structure-based drug design.
© 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
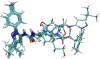
Similar articles
-
An Effective Reaction-Based Virtual Screening Method to Discover New CDK8 Ligands.ChemMedChem. 2025 May 5;20(9):e202400825. doi: 10.1002/cmdc.202400825. Epub 2025 Mar 10. ChemMedChem. 2025. PMID: 39972506
-
A molecular dynamics investigation of CDK8/CycC and ligand binding: conformational flexibility and implication in drug discovery.J Comput Aided Mol Des. 2018 Jun;32(6):671-685. doi: 10.1007/s10822-018-0120-3. Epub 2018 May 8. J Comput Aided Mol Des. 2018. PMID: 29737445 Free PMC article.
-
Highlighting the Major Role of Cyclin C in Cyclin-Dependent Kinase 8 Activity through Molecular Dynamics Simulations.Int J Mol Sci. 2024 May 15;25(10):5411. doi: 10.3390/ijms25105411. Int J Mol Sci. 2024. PMID: 38791449 Free PMC article.
-
Binding patterns and structure-activity relationship of CDK8 inhibitors.Bioorg Chem. 2020 Mar;96:103624. doi: 10.1016/j.bioorg.2020.103624. Epub 2020 Jan 25. Bioorg Chem. 2020. PMID: 32078847 Review.
-
Discovery and Development of Cyclin-Dependent Kinase 8 Inhibitors.Curr Med Chem. 2020;27(32):5429-5443. doi: 10.2174/0929867326666190402110528. Curr Med Chem. 2020. PMID: 30947649 Review.
Cited by
-
Super-Enhancers in the Regulation of Gene Transcription: General Aspects and Antitumor Targets.Acta Naturae. 2021 Jan-Mar;13(1):4-15. doi: 10.32607/actanaturae.11067. Acta Naturae. 2021. PMID: 33959383 Free PMC article.
-
Computation of host-guest binding free energies with a new quantum mechanics based mining minima algorithm.J Chem Phys. 2021 Mar 14;154(10):104122. doi: 10.1063/5.0040759. J Chem Phys. 2021. PMID: 33722015 Free PMC article.
-
Computation of Protein-Ligand Binding Free Energies with a Quantum Mechanics-Based Mining Minima Algorithm.J Chem Theory Comput. 2025 Apr 22;21(8):4236-4265. doi: 10.1021/acs.jctc.4c01707. Epub 2025 Apr 9. J Chem Theory Comput. 2025. PMID: 40202178 Free PMC article.
-
Pyrazolyl-Ureas as Interesting Scaffold in Medicinal Chemistry.Molecules. 2020 Jul 29;25(15):3457. doi: 10.3390/molecules25153457. Molecules. 2020. PMID: 32751358 Free PMC article. Review.
-
Transient States and Barriers from Molecular Simulations and the Milestoning Theory: Kinetics in Ligand-Protein Recognition and Compound Design.J Chem Theory Comput. 2020 Mar 10;16(3):1882-1895. doi: 10.1021/acs.jctc.9b01153. Epub 2020 Feb 20. J Chem Theory Comput. 2020. PMID: 32031801 Free PMC article.
References
-
- Tsutsui T, Fukasawa R, Tanaka A, Hirose Y, Ohkuma Y, Identification of target genes for the CDK subunits of the Mediator complex. Genes. Cells 2011, 16, 1208–1218. - PubMed
-
- Rickert P, Seghezzi W, Shanahan F, Cho H, Lees E, Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 1996, 12, 2631–2640. - PubMed

