Antagonists for Constitutively Active Mutant Estrogen Receptors: Insights into the Roles of Antiestrogen-Core and Side-Chain
- PMID: 30404440
- PMCID: PMC6469989
- DOI: 10.1021/acschembio.8b00877
Antagonists for Constitutively Active Mutant Estrogen Receptors: Insights into the Roles of Antiestrogen-Core and Side-Chain
Abstract
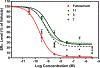
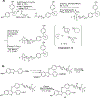
A major risk for patients having estrogen receptor α (ERα)-positive breast cancer is the recurrence of drug-resistant metastases after initial successful treatment with endocrine therapies. Recent studies have implicated a number of activating mutations in the ligand-binding domain of ERα that stabilize the agonist conformation as a prominent mechanism for this acquired resistance. There are several critical gaps in our knowledge regarding the specific pharmacophore requirements of an antagonist that could effectively inhibit all or most of the different mutant ERs. To address this, we screened various chemotypes for blocking mutant ER-mediated transcriptional signaling and identified RU58668 as a model compound that contains structural elements that support potent ligand-induced inhibition of mutant ERs. We designed and synthesized a focused library of novel antagonists and probed how small and large perturbations in different ligand structural regions influenced inhibitory activity on individual mutant ERs in breast cancer cells. Effective inhibition derives from both nonpolar and moderately polar motifs in a multifunctional side chain of the antagonists, with the nature of the ligand core making important contributions by increasing the potency of ligands possessing similar types of side chains. Some of our new antagonists potently blocked the transcriptional activity of the three most common mutant ERs (L536R, Y537S, D538G) and inhibited mutant ER-mediated cell proliferation. Supported by our molecular modeling, these studies provide new insights into the role of specific components, involving both the ligand core and multifunctional side chain, in suppressing wild-type and mutant ER-mediated transcription and breast cancer cell proliferation.
Figures



References
-
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, and Gustafsson JA (2007) Estrogen receptors: how do they signal and what are their targets, Physiol. Rev 87, 905–931. - PubMed
-
- Ariazi EA, Ariazi JL, Cordera F, and Jordan VC (2006) Estrogen receptors as therapeutic targets in breast cancer, Curr. Top. Med. Chem 6, 181–202. - PubMed
-
- Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, and Baum M (2008) Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial, Lancet Oncol. 9, 45–53. - PubMed
-
- Merenbakh-Lamin K, Ben-Baruch N, Yeheskel A, Dvir A, Soussan-Gutman L, Jeselsohn R, Yelensky R, Brown M, Miller VA, Sarid D, Rizel S, Klein B, Rubinek T, and Wolf I (2013) D538G mutation in estrogen receptor-alpha: A novel mechanism for acquired endocrine resistance in breast cancer, Cancer Res. 73, 6856–6864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

