Deletion of both the Tyrosine-Based Endocytosis Signal and the Endoplasmic Reticulum Retrieval Signal in the Cytoplasmic Tail of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Pigs
- PMID: 30404797
- PMCID: PMC6321913
- DOI: 10.1128/JVI.01758-18
Deletion of both the Tyrosine-Based Endocytosis Signal and the Endoplasmic Reticulum Retrieval Signal in the Cytoplasmic Tail of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Pigs
Abstract
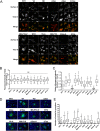
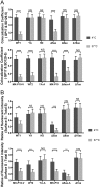
Porcine epidemic diarrhea virus (PEDV) causes high mortality in neonatal piglets. The PEDV spike (S) protein contains two intracellular sorting motifs, YxxΦ (tyrosine-based motif YEVF or YEAF) and KVHVQ at the cytoplasmic tail, yet their functions have not been fully elucidated. Some Vero cell-adapted and/or attenuated PEDV variants contain ablations in these two motifs. We hypothesized that these motifs contribute to viral pathogenicity. By transiently expressing PEDV S proteins with mutations in the motifs, we confirmed that the motif KVHVQ is involved in retention of the S proteins in the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC). In addition, we showed that the YxxΦ motif triggers endocytosis of S proteins. These two motifs synergistically regulate the level of S expressed on the cell surface. To investigate their role in viral pathogenicity, we generated three recombinant PEDVs by introducing deletions or a mutation in the two motifs of the infectious clone of PEDV PC22A strain (icPC22A): (i) icΔ10aa (ΔYxxΦEKVHVQ), (ii) icΔ5aa (ΔKVHVQ), and (iii) icYA (Y1378A, to an inactivated motif, AEVF). Infection of Vero cells with icΔ10aa resulted in larger syncytia and more virions, with reduced numbers of S protein projections on the surface compared with icPC22A. Furthermore, we orally inoculated five groups of 5-day-old gnotobiotic piglets with the three mutants, icPC22A, or a mock treatment. Mutant icΔ10aa caused less severe diarrhea rate and significantly milder intestinal lesions than icPC22A, icΔ5aa, and icYA. These data suggest that the deletion of both motifs can reduce the virulence of PEDV in piglets.IMPORTANCE Many coronaviruses (CoVs) possess conserved motifs YxxΦ and/or KxHxx/KKxx in the cytoplasmic tail of the S protein. The KxHxx/KKxx motif has been identified as the ER retrieval signal, but the function of the YxxΦ motif in the intracellular sorting of CoV S proteins remains controversial. In this study, we showed that the YxxΦ of PEDV S protein is an endocytosis signal. Furthermore, using reverse genetics technology, we evaluated its role in PEDV pathogenicity in neonatal piglets. Our results explain one attenuation mechanism of Vero cell-adapted PEDV variants lacking functional YxxΦ and KVHVQ motifs. Knowledge from this study may aid in the design of efficacious live attenuated vaccines against PEDV, as well as other CoVs bearing the same motif in their S protein.
Keywords: PEDV; YxxΦ; coronavirus; endocytosis; spike; virulence.
Copyright © 2019 American Society for Microbiology.
Figures






References
-
- Paarlberg PL. 2014. Updated estimated economic welfare impacts of porcine epidemic diarrhea virus (PEDV). http://ageconsearch.umn.edu/bitstream/174517/2/14-4.Updated%20Estimated%....
-
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. doi: 10.1177/1040638713501675. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

