Sensitivity to Broadly Neutralizing Antibodies of Recently Transmitted HIV-1 Clade CRF02_AG Viruses with a Focus on Evolution over Time
- PMID: 30404804
- PMCID: PMC6321924
- DOI: 10.1128/JVI.01492-18
Sensitivity to Broadly Neutralizing Antibodies of Recently Transmitted HIV-1 Clade CRF02_AG Viruses with a Focus on Evolution over Time
Abstract
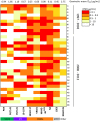
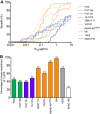
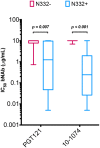
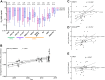
Broadly neutralizing antibodies (bnAbs) are promising agents for prevention and/or treatment of HIV-1 infection. However, the diversity among HIV-1 envelope (Env) glycoproteins impacts bnAb potency and breadth. Neutralization data on the CRF02_AG clade are scarce although it is highly prevalent in West Africa and Europe. We assessed the sensitivity to bnAbs of a panel of 33 early transmitted CRF02_AG viruses over a 15-year period of the French epidemic (1997 to 2012). Env pseudotyped CRF02_AG viruses were best neutralized by the CD4 binding site (CD4bs)-directed bnAbs (VRC01, 3BNC117, NIH45-46G54W, and N6) and the gp41 membrane-proximal external region (MPER)-directed bnAb 10E8 in terms of both potency and breadth. We observed a higher resistance to bnAbs targeting the V1V2-glycan region (PG9 and PGT145) and the V3-glycan region (PGT121 and 10-1074). Combinations were required to achieve full coverage across this subtype. We observed increased resistance to bnAbs targeting the CD4bs linked to the diversification of CRF02_AG Env over the course of the epidemic, a phenomenon which was previously described for subtypes B and C. These data on the sensitivity to bnAbs of CRF02_AG viruses, including only recently transmitted viruses, will inform future passive immunization studies. Considering the drift of the HIV-1 species toward higher resistance to neutralizing antibodies, it appears necessary to keep updating existing panels for evaluation of future vaccine and passive immunization studies.IMPORTANCE Major progress occurred during the last decade leading to the isolation of human monoclonal antibodies, termed broadly neutralizing antibodies (bnAbs) due to their capacity to neutralize various strains of HIV-1. Several clinical trials are under way in order to evaluate their efficacy in preventive or therapeutic strategies. However, no single bnAb is active against 100% of strains. It is important to gather data on the sensitivity to neutralizing antibodies of all genotypes, especially those more widespread in regions where the prevalence of HIV-1 infection is high. Here, we assembled a large panel of clade CRF02_AG viruses, the most frequent genotype circulating in West Africa and the second most frequent found in several European countries. We evaluated their sensitivities to bnAbs, including those most advanced in clinical trials, and looked for the best combinations. In addition, we observed a trend toward increased resistance to bnAbs over the course of the epidemic.
Keywords: evolution; human immunodeficiency virus; neutralizing antibodies.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Evaluation of susceptibility of HIV-1 CRF01_AE variants to neutralization by a panel of broadly neutralizing antibodies.Arch Virol. 2018 Dec;163(12):3303-3315. doi: 10.1007/s00705-018-4011-7. Epub 2018 Sep 8. Arch Virol. 2018. PMID: 30196320
-
Distinct region-specific neutralization profiles of contemporary HIV-1 clade C against best-in-class broadly neutralizing antibodies.J Virol. 2025 Jun 17;99(6):e0000825. doi: 10.1128/jvi.00008-25. Epub 2025 May 16. J Virol. 2025. PMID: 40377318 Free PMC article.
-
Neutralizing Activity of Broadly Neutralizing Anti-HIV-1 Antibodies against Clade B Clinical Isolates Produced in Peripheral Blood Mononuclear Cells.J Virol. 2018 Feb 12;92(5):e01883-17. doi: 10.1128/JVI.01883-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237833 Free PMC article.
-
Strategies for induction of HIV-1 envelope-reactive broadly neutralizing antibodies.J Int AIDS Soc. 2021 Nov;24 Suppl 7(Suppl 7):e25831. doi: 10.1002/jia2.25831. J Int AIDS Soc. 2021. PMID: 34806332 Free PMC article. Review.
-
Broadly neutralizing antibodies for the treatment and prevention of HIV infection.Curr Opin HIV AIDS. 2020 Jan;15(1):49-55. doi: 10.1097/COH.0000000000000600. Curr Opin HIV AIDS. 2020. PMID: 31764199 Free PMC article. Review.
Cited by
-
Phenotypic Characterization of Subtype A and Recombinant AC Transmitted/Founder Viruses from a Rwandan HIV-1 Heterosexual Transmission Cohort.Viruses. 2024 Oct 30;16(11):1706. doi: 10.3390/v16111706. Viruses. 2024. PMID: 39599821 Free PMC article.
-
HIV-1 Entry and Prospects for Protecting against Infection.Microorganisms. 2021 Jan 22;9(2):228. doi: 10.3390/microorganisms9020228. Microorganisms. 2021. PMID: 33499233 Free PMC article. Review.
-
Differences in neutralizing antibody sensitivities and envelope characteristics indicate distinct antigenic properties of Nigerian HIV-1 subtype G and CRF02_AG.Virol J. 2024 Jun 29;21(1):148. doi: 10.1186/s12985-024-02394-y. Virol J. 2024. PMID: 38951814 Free PMC article.
-
Impact of HIV-1 Diversity on Its Sensitivity to Neutralization.Vaccines (Basel). 2019 Jul 25;7(3):74. doi: 10.3390/vaccines7030074. Vaccines (Basel). 2019. PMID: 31349655 Free PMC article. Review.
-
Construction and Characterization of HIV-1 env-Pseudoviruses of the Recombinant Form CRF63_02A and Subtype A6.Bull Exp Biol Med. 2022 Apr;172(6):729-733. doi: 10.1007/s10517-022-05466-7. Epub 2022 May 2. Bull Exp Biol Med. 2022. PMID: 35501651
References
-
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators . 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi:10.1056/NEJMoa0908492. - DOI - PubMed
-
- Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao H-X, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi:10.1056/NEJMoa1113425. - DOI - PMC - PubMed
-
- Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Kwon YD, Scheid J, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817. doi:10.1126/science.1192819. - DOI - PMC - PubMed
-
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Klein F, Gao H, Gnanapragasam PNP, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. 2011. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science 334:1289–1293. doi:10.1126/science.1213782. - DOI - PMC - PubMed
-
- Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi:10.1126/science.1187659. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

