Development of a Primary Human Cell Model for the Study of Human Cytomegalovirus Replication and Spread within Salivary Epithelium
- PMID: 30404806
- PMCID: PMC6340043
- DOI: 10.1128/JVI.01608-18
Development of a Primary Human Cell Model for the Study of Human Cytomegalovirus Replication and Spread within Salivary Epithelium
Abstract
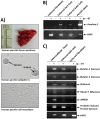
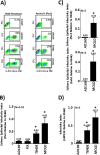
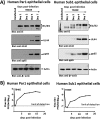
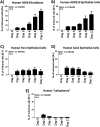
Various aspects of human cytomegalovirus (HCMV) pathogenesis, including its ability to replicate in specific cells and tissues and the mechanism(s) of horizontal transmission, are not well understood, predominantly because of the strict species specificity exhibited by HCMV. Murine CMV (MCMV), which contains numerous gene segments highly similar to those of HCMV, has been useful for modeling some aspects of CMV pathogenesis; however, it remains essential to build relevant human cell-based systems to investigate how the HCMV counterparts function. The salivary gland epithelium is a site of persistence for both human and murine cytomegaloviruses, and salivary secretions appear to play an important role in horizontal transmission. Therefore, it is important to understand how HCMV is replicating within the glandular epithelial cells so that it might be possible to therapeutically prevent transmission. In the present study, we describe the development of a salivary epithelial model derived from primary human "salispheres." Initial infection of these primary salivary cells with HCMV occurs in a manner similar to that reported for established epithelial lines, in that gH/gL/UL128/UL130/UL131A (pentamer)-positive strains can infect and replicate, while laboratory-adapted pentamer-null strains do not. However, while HCMV enters the lytic phase and produces virus in salivary epithelial cells, it fails to exhibit robust spread throughout the culture and persists in a low percentage of salivary cells. The present study demonstrates the utility of these primary tissue-derived cells for studying HCMV replication in salivary epithelial cells in vitroIMPORTANCE Human cytomegalovirus (HCMV) infects the majority of the world's population, and although it typically establishes a quiescent infection with little to no disease in most individuals, the virus is responsible for a variety of devastating sequelae in immunocompromised adults and in developing fetuses. Therefore, identifying the viral properties essential for replication, spread, and horizontal transmission is an important area of medical science. Our studies use novel human salivary gland-derived cellular models to investigate the molecular details by which HCMV replicates in salivary epithelial cells and provide insight into the mechanisms by which the virus persists in the salivary epithelium, where it gains access to fluids centrally important for horizontal transmission.
Keywords: HCMV; cytomegalovirus; horizontal dissemination; latency; lytic replication; organoid; salisphere; salivary gland.
Copyright © 2019 American Society for Microbiology.
Figures
Similar articles
-
Loss of the Human Cytomegalovirus US16 Protein Abrogates Virus Entry into Endothelial and Epithelial Cells by Reducing the Virion Content of the Pentamer.J Virol. 2017 May 12;91(11):e00205-17. doi: 10.1128/JVI.00205-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28331097 Free PMC article.
-
CD147 Promotes Entry of Pentamer-Expressing Human Cytomegalovirus into Epithelial and Endothelial Cells.mBio. 2018 May 8;9(3):e00781-18. doi: 10.1128/mBio.00781-18. mBio. 2018. PMID: 29739904 Free PMC article.
-
The M33 G protein-coupled receptor encoded by murine cytomegalovirus is dispensable for hematogenous dissemination but is required for growth within the salivary gland.J Virol. 2014 Oct;88(20):11811-24. doi: 10.1128/JVI.01006-14. Epub 2014 Aug 6. J Virol. 2014. PMID: 25100846 Free PMC article.
-
Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications.Rev Med Virol. 2010 May;20(3):136-55. doi: 10.1002/rmv.645. Rev Med Virol. 2010. PMID: 20084641 Review.
-
Endothelial cells in human cytomegalovirus infection: one host cell out of many or a crucial target for virus spread?Thromb Haemost. 2009 Dec;102(6):1057-63. doi: 10.1160/TH09-04-0213. Thromb Haemost. 2009. PMID: 19967135 Review.
Cited by
-
Human Nasal Turbinate Tissues in Organ Culture as a Model for Human Cytomegalovirus Infection at the Mucosal Entry Site.J Virol. 2020 Sep 15;94(19):e01258-20. doi: 10.1128/JVI.01258-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32727881 Free PMC article.
-
The Detection of CMV in Saliva Can Mark a Systemic Infection with CMV in Renal Transplant Recipients.Int J Mol Sci. 2019 Oct 22;20(20):5230. doi: 10.3390/ijms20205230. Int J Mol Sci. 2019. PMID: 31652514 Free PMC article.
-
Correspondence Between Cytomegalovirus Immunoglobulin-G Levels Measured in Saliva and Serum.Front Immunol. 2020 Aug 28;11:2095. doi: 10.3389/fimmu.2020.02095. eCollection 2020. Front Immunol. 2020. PMID: 32983163 Free PMC article. Clinical Trial.
-
Engineering a Model to Study Viral Infections: Bioprinting, Microfluidics, and Organoids to Defeat Coronavirus Disease 2019 (COVID-19).Int J Bioprint. 2020 Aug 28;6(4):302. doi: 10.18063/ijb.v6i4.302. eCollection 2020. Int J Bioprint. 2020. PMID: 33089000 Free PMC article. Review.
-
Rescue of Pentamer-Null Strains of Human Cytomegalovirus in Epithelial Cells by Use of Histone Deacetylase Inhibitors Reveals an Additional Postentry Function for the Pentamer Complex.J Virol. 2022 Apr 27;96(8):e0003122. doi: 10.1128/jvi.00031-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343807 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2702–2772. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol2 Lippincott Williams & Williams, Philadelphia, PA.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

