An interaction network between the SNARE VAMP7 and Rab GTPases within a ciliary membrane-targeting complex
- PMID: 30404838
- PMCID: PMC6307879
- DOI: 10.1242/jcs.222034
An interaction network between the SNARE VAMP7 and Rab GTPases within a ciliary membrane-targeting complex
Abstract
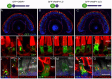
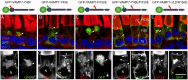
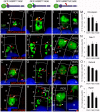
The Arf4-rhodopsin complex (mediated by the VxPx motif in rhodopsin) initiates expansion of vertebrate rod photoreceptor cilia-derived light-sensing organelles through stepwise assembly of a conserved trafficking network. Here, we examine its role in the sorting of VAMP7 (also known as TI-VAMP) - an R-SNARE possessing a regulatory longin domain (LD) - into rhodopsin transport carriers (RTCs). During RTC formation and trafficking, VAMP7 colocalizes with the ciliary cargo rhodopsin and interacts with the Rab11-Rabin8-Rab8 trafficking module. Rab11 and Rab8 bind the VAMP7 LD, whereas Rabin8 (also known as RAB3IP) interacts with the SNARE domain. The Arf/Rab11 effector FIP3 (also known as RAB11FIP3) regulates VAMP7 access to Rab11. At the ciliary base, VAMP7 forms a complex with the cognate SNAREs syntaxin 3 and SNAP-25. When expressed in transgenic animals, a GFP-VAMP7ΔLD fusion protein and a Y45E phosphomimetic mutant colocalize with endogenous VAMP7. The GFP-VAMP7-R150E mutant displays considerable localization defects that imply an important role of the R-SNARE motif in intracellular trafficking, rather than cognate SNARE pairing. Our study defines the link between VAMP7 and the ciliary targeting nexus that is conserved across diverse cell types, and contributes to general understanding of how functional Arf and Rab networks assemble SNAREs in membrane trafficking.
Keywords: Cilium; Rab GTPase; Rhodopsin; SNARE; Sensory receptor; VAMP7.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




Similar articles
-
The ins and outs of the Arf4-based ciliary membrane-targeting complex.Small GTPases. 2021 Jan;12(1):1-12. doi: 10.1080/21541248.2019.1616355. Epub 2019 May 17. Small GTPases. 2021. PMID: 31068062 Free PMC article. Review.
-
The Arf and Rab11 effector FIP3 acts synergistically with ASAP1 to direct Rabin8 in ciliary receptor targeting.J Cell Sci. 2015 Apr 1;128(7):1375-85. doi: 10.1242/jcs.162925. Epub 2015 Feb 11. J Cell Sci. 2015. PMID: 25673879 Free PMC article.
-
Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4.EMBO J. 2009 Feb 4;28(3):183-92. doi: 10.1038/emboj.2008.267. Epub 2009 Jan 15. EMBO J. 2009. PMID: 19153612 Free PMC article.
-
Rabin8 phosphorylated by NDR2, the canine early retinal degeneration gene product, directs rhodopsin Golgi-to-cilia trafficking.J Cell Sci. 2025 Jan 15;138(2):JCS263401. doi: 10.1242/jcs.263401. Epub 2025 Jan 23. J Cell Sci. 2025. PMID: 39774853 Free PMC article.
-
Molecular assemblies that control rhodopsin transport to the cilia.Vision Res. 2012 Dec 15;75:5-10. doi: 10.1016/j.visres.2012.07.015. Epub 2012 Aug 7. Vision Res. 2012. PMID: 22892112 Free PMC article. Review.
Cited by
-
The ins and outs of the Arf4-based ciliary membrane-targeting complex.Small GTPases. 2021 Jan;12(1):1-12. doi: 10.1080/21541248.2019.1616355. Epub 2019 May 17. Small GTPases. 2021. PMID: 31068062 Free PMC article. Review.
-
Autophagy Drives Galectin-1 Secretion From Tumor-Associated Macrophages Facilitating Hepatocellular Carcinoma Progression.Front Cell Dev Biol. 2021 Sep 6;9:741820. doi: 10.3389/fcell.2021.741820. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34552935 Free PMC article.
-
Syntaxin 3 is essential for photoreceptor outer segment protein trafficking and survival.Proc Natl Acad Sci U S A. 2020 Aug 25;117(34):20615-20624. doi: 10.1073/pnas.2010751117. Epub 2020 Aug 10. Proc Natl Acad Sci U S A. 2020. PMID: 32778589 Free PMC article.
-
Syntaxin 3B: A SNARE Protein Required for Vision.Int J Mol Sci. 2024 Oct 3;25(19):10665. doi: 10.3390/ijms251910665. Int J Mol Sci. 2024. PMID: 39408994 Free PMC article. Review.
-
Proliferative signaling by ERBB proteins and RAF/MEK/ERK effectors in polycystic kidney disease.Cell Signal. 2020 Mar;67:109497. doi: 10.1016/j.cellsig.2019.109497. Epub 2019 Dec 9. Cell Signal. 2020. PMID: 31830556 Free PMC article. Review.
References
-
- Bachmann-Gagescu R., Phelps I. G., Stearns G., Link B. A., Brockerhoff S. E., Moens C. B. and Doherty D. (2011). The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Hum. Mol. Genet. 20, 4041-4055. 10.1093/hmg/ddr332 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

