Assembly of the β4-Integrin Interactome Based on Proximal Biotinylation in the Presence and Absence of Heterodimerization
- PMID: 30404858
- PMCID: PMC6356083
- DOI: 10.1074/mcp.RA118.001095
Assembly of the β4-Integrin Interactome Based on Proximal Biotinylation in the Presence and Absence of Heterodimerization
Abstract
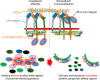
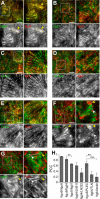
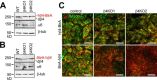
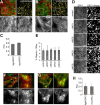
Integrin-mediated laminin adhesions mediate epithelial cell anchorage to basement membranes and are critical regulators of epithelial cell polarity. Integrins assemble large multiprotein complexes that link to the cytoskeleton and convey signals into the cells. Comprehensive proteomic analyses of actin network-linked focal adhesions (FA) have been performed, but the molecular composition of intermediate filament-linked hemidesmosomes (HD) remains incompletely characterized. Here we have used proximity-dependent biotin identification (BioID) technology to label and characterize the interactome of epithelia-specific β4-integrin that, as α6β4-heterodimer, forms the core of HDs. The analysis identified ∼150 proteins that were specifically labeled by BirA-tagged integrin-β4. In addition to known HDs proteins, the interactome revealed proteins that may indirectly link integrin-β4 to actin-connected protein complexes, such as FAs and dystrophin/dystroglycan complexes. The specificity of the screening approach was validated by confirming the HD localization of two candidate β4-interacting proteins, utrophin (UTRN) and ELKS/Rab6-interacting/CAST family member 1 (ERC1). Interestingly, although establishment of functional HDs depends on the formation of α6β4-heterodimers, the assembly of β4-interactome was not strictly dependent on α6-integrin expression. Our survey to the HD interactome sets a precedent for future studies and provides novel insight into the mechanisms of HD assembly and function of the β4-integrin.
Keywords: BioID; Cell Adhesion; Epithelium; Extracellular Matrix; Hemidesmosome; Integrin; Kidney Function or Biology; Mass Spectrometry; Protein Complex Analysis.
© 2019 Myllymäki et al.
Conflict of interest statement
We declare that they have no conflicts of interest with the contents of this article
Figures





Similar articles
-
Comparative interactomics analysis reveals potential regulators of α6β4 distribution in keratinocytes.Biol Open. 2020 Aug 13;9(8):bio054155. doi: 10.1242/bio.054155. Biol Open. 2020. PMID: 32709696 Free PMC article.
-
Loss of α6β4 Integrin-Mediated Hemidesmosomes Promotes Prostate Epithelial Cell Migration by Stimulating Focal Adhesion Dynamics.Front Cell Dev Biol. 2022 Jul 7;10:886569. doi: 10.3389/fcell.2022.886569. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35874837 Free PMC article.
-
Crucial role of the specificity-determining loop of the integrin beta4 subunit in the binding of cells to laminin-5 and outside-in signal transduction.J Biol Chem. 2003 Oct 3;278(40):38707-14. doi: 10.1074/jbc.M301637200. Epub 2003 Jul 16. J Biol Chem. 2003. PMID: 12867433
-
Advances and perspectives of the architecture of hemidesmosomes: lessons from structural biology.Cell Adh Migr. 2009 Oct-Dec;3(4):361-4. doi: 10.4161/cam.3.4.9525. Epub 2009 Oct 16. Cell Adh Migr. 2009. PMID: 19736524 Free PMC article. Review.
-
Integrin beta4, keratinocytes and papillomavirus infection.Int J Mol Med. 2006 Feb;17(2):195-202. Int J Mol Med. 2006. PMID: 16391815 Review.
Cited by
-
Junctional epithelium and hemidesmosomes: Tape and rivets for solving the "percutaneous device dilemma" in dental and other permanent implants.Bioact Mater. 2022 Mar 19;18:178-198. doi: 10.1016/j.bioactmat.2022.03.019. eCollection 2022 Dec. Bioact Mater. 2022. PMID: 35387164 Free PMC article. Review.
-
The extracellular matrix component perlecan/HSPG2 regulates radioresistance in prostate cancer cells.Front Cell Dev Biol. 2024 Aug 1;12:1452463. doi: 10.3389/fcell.2024.1452463. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39149513 Free PMC article.
-
Phagocytosis by the retinal pigment epithelium: New insights into polarized cell mechanics.Bioessays. 2025 Jan;47(1):e2300197. doi: 10.1002/bies.202300197. Epub 2024 Dec 11. Bioessays. 2025. PMID: 39663766 Free PMC article. Review.
-
β2 Integrin Signaling Cascade in Neutrophils: More Than a Single Function.Front Immunol. 2021 Feb 18;11:619925. doi: 10.3389/fimmu.2020.619925. eCollection 2020. Front Immunol. 2021. PMID: 33679708 Free PMC article. Review.
-
Regulation of Kinase Signaling Pathways by α6β4-Integrins and Plectin in Prostate Cancer.Cancers (Basel). 2022 Dec 27;15(1):149. doi: 10.3390/cancers15010149. Cancers (Basel). 2022. PMID: 36612146 Free PMC article. Review.
References
-
- O'Brien L. E., Jou T. S., Pollack A. L., Zhang Q., Hansen S. H., Yurchenco P., and Mostov K. E. (2001) Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat. Cell Biol. 3, 831–838 - PubMed
-
- Geiger T., and Zaidel-Bar R. (2012) Opening the floodgates: proteomics and the integrin adhesome. Curr. Opin. Cell Biol. 24, 562–568 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

