The geographic mosaic of coevolution in mutualistic networks
- PMID: 30404910
- PMCID: PMC6255164
- DOI: 10.1073/pnas.1809088115
The geographic mosaic of coevolution in mutualistic networks
Abstract
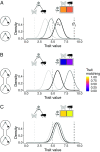
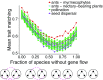
Ecological interactions shape adaptations through coevolution not only between pairs of species but also through entire multispecies assemblages. Local coevolution can then be further altered through spatial processes that have been formally partitioned in the geographic mosaic theory of coevolution. A major current challenge is to understand the spatial patterns of coadaptation that emerge across ecosystems through the interplay between gene flow and selection in networks of interacting species. Here, we combine a coevolutionary model, network theory, and empirical information on species interactions to investigate how gene flow and geographical variation in selection affect trait patterns in mutualistic networks. We show that gene flow has the surprising effect of favoring trait matching, especially among generalist species in species-rich networks typical of pollination and seed dispersal interactions. Using an analytical approximation of our model, we demonstrate that gene flow promotes trait matching by making the adaptive landscapes of different species more similar to each other. We use this result to show that the progressive loss of gene flow associated with habitat fragmentation may undermine coadaptation in mutualisms. Our results therefore provide predictions of how spatial processes shape the evolution of species-rich interactions and how the widespread fragmentation of natural landscapes may modify the coevolutionary process.
Keywords: coadaptation; ecological networks; gene flow; mutualism; trait matching.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Thompson JN. The Geographic Mosaic of Coevolution. Univ of Chicago Press; Chicago: 2005.
-
- Pauw A, Stofberg J, Waterman RJ. Flies and flowers in Darwin’s race. Evolution. 2009;63:268–279. - PubMed
-
- McCann KS. Food Webs. Princeton Univ Press; Princeton: 2011.
-
- Bascompte J, Jordano P. Mutualistic Networks. Princeton Univ Press; Princeton: 2014.

