Dynamic recognition and linkage specificity in K63 di-ubiquitin and TAB2 NZF domain complex
- PMID: 30405169
- PMCID: PMC6220233
- DOI: 10.1038/s41598-018-34605-2
Dynamic recognition and linkage specificity in K63 di-ubiquitin and TAB2 NZF domain complex
Abstract
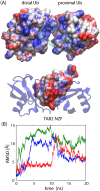
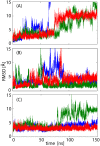
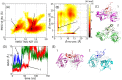
Poly-ubiquitin (poly-Ub) is involved in various cellular processes through the linkage-specific recognition of Ub-binding domains (UBD). In this study, using molecular dynamics (MD) simulation together with an enhanced sampling method, we demonstrated that K63-linked di-Ub recognizes the NZF domain of TAB2, a zinc finger UBD, in an ensemble of highly dynamic structures that form from the weak interactions between UBD and the flexible linker connecting the two Ubs. However, the K63 di-Ub/TAB2 NZF complex showed a much more compact and stable ensemble than the non-native complexes, linear di-Ub/TAB2 NZF and K33 di-Ub/TAB2 NZF, that were modeled from linear di-Ub/HOIL-1L NZF and K33 di-Ub/TRABID NZF1, respectively. We further demonstrated the importance of the length and position of the Ub-Ub linker in the results of MD simulations of K63 di-Ub/TAB2 NZF by changing the Ub linkage from the native K63 to four different non-native linkages, linear, K6, K11, and K48, while maintaining inter-molecular contacts in the native complex. No systems with non-native linkage maintained the native binding configuration. These simulation results provide an atomistic picture of the linkage specific recognition of poly-Ubs leading to the biological functions such as cellular colocalization of various component proteins in the signal transduction pathways.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Structural basis for specific recognition of K6-linked polyubiquitin chains by the TAB2 NZF domain.Biophys J. 2021 Aug 17;120(16):3355-3362. doi: 10.1016/j.bpj.2021.06.037. Epub 2021 Jul 7. Biophys J. 2021. PMID: 34242591 Free PMC article.
-
Solution structure of the HOIL-1L NZF domain reveals a conformational switch regulating linear ubiquitin affinity.J Biol Chem. 2023 Sep;299(9):105165. doi: 10.1016/j.jbc.2023.105165. Epub 2023 Aug 16. J Biol Chem. 2023. PMID: 37595872 Free PMC article.
-
Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3.EMBO J. 2009 Dec 16;28(24):3903-9. doi: 10.1038/emboj.2009.345. EMBO J. 2009. PMID: 19927120 Free PMC article.
-
Ubiquitin-binding domains.Biochem J. 2006 Nov 1;399(3):361-72. doi: 10.1042/BJ20061138. Biochem J. 2006. PMID: 17034365 Free PMC article. Review.
-
Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series.EMBO Rep. 2008 Jun;9(6):536-42. doi: 10.1038/embor.2008.93. EMBO Rep. 2008. PMID: 18516089 Free PMC article. Review.
Cited by
-
Ubiquitination drives COPI priming and Golgi SNARE localization.Elife. 2022 Jul 29;11:e80911. doi: 10.7554/eLife.80911. Elife. 2022. PMID: 35904239 Free PMC article.
-
Exploring Configuration Space and Path Space of Biomolecules Using Enhanced Sampling Techniques-Searching for Mechanism and Kinetics of Biomolecular Functions.Int J Mol Sci. 2018 Oct 15;19(10):3177. doi: 10.3390/ijms19103177. Int J Mol Sci. 2018. PMID: 30326661 Free PMC article.
-
Multiscale Enhanced Sampling Using Machine Learning.Life (Basel). 2021 Oct 12;11(10):1076. doi: 10.3390/life11101076. Life (Basel). 2021. PMID: 34685447 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

