The Endophytic Mycobiome of European Ash and Sycamore Maple Leaves - Geographic Patterns, Host Specificity and Influence of Ash Dieback
- PMID: 30405540
- PMCID: PMC6207852
- DOI: 10.3389/fmicb.2018.02345
The Endophytic Mycobiome of European Ash and Sycamore Maple Leaves - Geographic Patterns, Host Specificity and Influence of Ash Dieback
Abstract
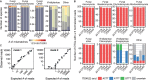
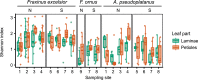
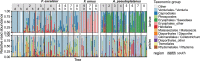
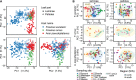
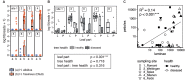
The European ash (Fraxinus excelsior) is threatened by the introduced ascomycete Hymenoscyphus fraxineus, the causal agent of ash dieback. Endophytic fungi are known to modulate their host's resistance against pathogens. To understand possible consequences of ash dieback on the endophytic mycobiome, F. excelsior leaves were collected in naturally regenerated forests and the fungal communities analyzed by classic culture and Illumina amplicon sequencing using a newly developed and validated fungal-specific primer. Collections were done in the area infested by ash dieback north of the Alps, and in the disease free area on the south side. Sycamore maple (Acer pseudoplatanus) was additionally collected, as well as the flowering ash (F. ornus), which occurs naturally in the south and shows tolerance to ash dieback. Both cultivation and amplicon sequencing revealed characteristic endophytic fungal communities dominated by several strictly host specific Venturia species. On A. pseudoplatanus, a hitherto undescribed Venturia species was identified. Due to its dominance on F. excelsior, V. fraxini is unlikely to go extinct in case of reduced host densities. A majority of species was not strictly host specific and is therefore likely less affected by ash dieback in the future. Still, shifts in community structure and loss of genetic diversity cannot be excluded. The potentially endangered endophyte Hymenoscyphus albidus was rarely found. In addition to host specificity, species with preferences for leaf laminae or petioles were found. We also detected considerable geographical variation between sampling sites and clear differences between the two sides of the Alps for endophytes of F. excelsior, but not A. pseudoplatanus. Since sycamore maple is not affected by an epidemic, this could point toward an influence of ash dieback on ash communities, although firm conclusions are not possible because of host preferences and climatic differences. Furthermore, the mycobiota of F. excelsior trees with or without dieback symptoms were compared, but no clear differences were detected. Besides methodical refinement, our study provides comprehensive data on the ash mycobiome that we expect to be subject to changes caused by an emerging disease of the host tree.
Keywords: ash dieback; cryptic extinction; emerging disease; endophytic fungi; fungal-specific primers; invasive pathogen; mock community.
Figures
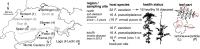



References
-
- Abegg M., Brändli U. B., Cioldi F., Fischer C., Herold-Bonardi A., Huber M., et al. (2014). Viertes Schweizerisches Landesforstinventar–Ergebnistabellen und Karten im Internet zum LFI 2009-2013 (LFI4b). Available at: http://www.lfi.ch/resultate/
-
- Bakys R., Vasaitis R., Barklund P., Ihrmark K., Stenlid J. (2009a). Investigations concerning the role of Chalara fraxinea in declining Fraxinus excelsior. Plant Pathol. 58 284–292. 10.1111/j.1365-3059.2008.01977.x - DOI
-
- Bakys R., Vasaitis R., Barklund P., Thomsen I. M., Stenlid J. (2009b). Occurrence and pathogenicity of fungi in necrotic and non-symptomatic shoots of declining common ash (Fraxinus excelsior) in Sweden. Eur. J. For. Res. 128 51–60. 10.1007/s10342-008-0238-2 - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

