Antimicrobial Resistance Profile of mcr-1 Positive Clinical Isolates of Escherichia coli in China From 2013 to 2016
- PMID: 30405572
- PMCID: PMC6206212
- DOI: 10.3389/fmicb.2018.02514
Antimicrobial Resistance Profile of mcr-1 Positive Clinical Isolates of Escherichia coli in China From 2013 to 2016
Abstract
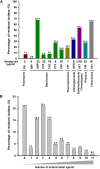
Multidrug-resistant (MDR) Escherichia coli poses a great challenge for public health in recent decades. Polymyxins have been reconsidered as a valuable therapeutic option for the treatment of infections caused by MDR E. coli. A plasmid-encoded colistin resistance gene mcr-1 encoding phosphoethanolamine transferase has been recently described in Enterobacteriaceae. In this study, a total of 123 E. coli isolates obtained from patients with diarrheal diseases in China were used for the genetic analysis of colistin resistance in clinical isolates. Antimicrobial resistance profile of polymyxin B (PB) and 11 commonly used antimicrobial agents were determined. Among the 123 E. coli isolates, 9 isolates (7.3%) were resistant to PB and PCR screening showed that seven (5.7%) isolates carried the mcr-1 gene. A hybrid sequencing analysis using single-molecule, real-time (SMRT) sequencing and Illumina sequencing was then performed to resolve the genomes of the seven mcr-1 positive isolates. These seven isolates harbored multiple plasmids and are MDR, with six isolates carrying one mcr-1 positive plasmid and one isolate (14EC033) carrying two mcr-1 positive plasmids. These eight mcr-1 positive plasmids belonged to the IncX4, IncI2, and IncP1 types. In addition, the mcr-1 gene was the solo antibiotic resistance gene identified in the mcr-1 positive plasmids, while the rest of the antibiotic resistance genes were mostly clustered into one or two plasmids. Interestingly, one mcr-1 positive isolate (14EC047) was susceptible to PB, and we showed that the activity of MCR-1-mediated colistin resistance was not phenotypically expressed in 14EC047 host strain. Furthermore, three isolates exhibited resistance to PB but did not carry previously reported mcr-related genes. Multilocus sequence typing (MLST) showed that these mcr-1 positive E. coli isolates belonged to five different STs, and three isolates belonged to ST301 which carried multiple virulence factors related to diarrhea. Additionally, the mcr-1 positive isolates were all susceptible to imipenem (IMP), suggesting that IMP could be used to treat infection caused by mcr-1 positive E. coli isolates. Collectively, this study showed a high occurrence of mcr-1 positive plasmids in patients with diarrheal diseases of Guangzhou in China and the abolishment of the MCR-1 mediated colistin resistance in one E. coli isolate.
Keywords: Escherichia coli; clinical isolates; mcr-1; multidrug-resistant; plasmid.
Figures


References
-
- Afset J. E., Bruant G., Brousseau R., Harel J., Anderssen E., Bevanger L., et al. (2006). Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J. Clin. Microbiol. 44 3703–3711. 10.1128/Jcm.00429-06 - DOI - PMC - PubMed
-
- Bai L., Hurley D., Li J., Meng Q., Wang J., Fanning S., et al. (2016). Characterisation of multidrug-resistant Shiga toxin-producing Escherichia coli cultured from pigs in China: co-occurrence of extended-spectrum beta-lactamase- and mcr-1-encoding genes on plasmids. Int. J. Antimicrob. Agents 48 445–448. 10.1016/j.ijantimicag.2016.06.021 - DOI - PubMed
LinkOut - more resources
Full Text Sources

