Microbiomes Associated With Foods From Plant and Animal Sources
- PMID: 30405589
- PMCID: PMC6206262
- DOI: 10.3389/fmicb.2018.02540
Microbiomes Associated With Foods From Plant and Animal Sources
Abstract
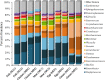
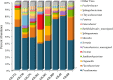
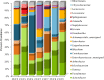
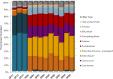
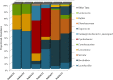
Food microbiome composition impacts food safety and quality. The resident microbiota of many food products is influenced throughout the farm to fork continuum by farming practices, environmental factors, and food manufacturing and processing procedures. Currently, most food microbiology studies rely on culture-dependent methods to identify bacteria. However, advances in high-throughput DNA sequencing technologies have enabled the use of targeted 16S rRNA gene sequencing to profile complex microbial communities including non-culturable members. In this study we used 16S rRNA gene sequencing to assess the microbiome profiles of plant and animal derived foods collected at two points in the manufacturing process; post-harvest/pre-retail (cilantro) and retail (cilantro, masala spice mixes, cucumbers, mung bean sprouts, and smoked salmon). Our findings revealed microbiome profiles, unique to each food, that were influenced by the moisture content (dry spices, fresh produce), packaging methods, such as modified atmospheric packaging (mung bean sprouts and smoked salmon), and manufacturing stage (cilantro prior to retail and at retail). The masala spice mixes and cucumbers were comprised mainly of Proteobacteria, Firmicutes, and Actinobacteria. Cilantro microbiome profiles consisted mainly of Proteobacteria, followed by Bacteroidetes, and low levels of Firmicutes and Actinobacteria. The two brands of mung bean sprouts and the three smoked salmon samples differed from one another in their microbiome composition, each predominated by either by Firmicutes or Proteobacteria. These data demonstrate diverse and highly variable resident microbial communities across food products, which is informative in the context of food safety, and spoilage where indigenous bacteria could hamper pathogen detection, and limit shelf life.
Keywords: 16S rRna; food; metagenomics; microbiome; produce; seafood; spices.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources

