Real Time Monitoring of NADPH Concentrations in Corynebacterium glutamicum and Escherichia coli via the Genetically Encoded Sensor mBFP
- PMID: 30405597
- PMCID: PMC6207642
- DOI: 10.3389/fmicb.2018.02564
Real Time Monitoring of NADPH Concentrations in Corynebacterium glutamicum and Escherichia coli via the Genetically Encoded Sensor mBFP
Abstract
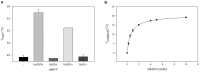
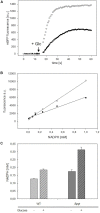
Analyses of intracellular NADPH concentrations are prerequisites for the design of microbial production strains and process optimization. mBFP was described as metagenomics derived, blue fluorescent protein showing NADPH-dependent fluorescence. Characterization of mBFP showed a high specificity for binding of NADPH (K D 0.64 mM) and no binding of NADH, the protein exclusively amplified fluorescence of NADPH. mBFP catalyzed the NADPH-dependent reduction of benzaldehyde and further aldehydes, which fits to its classification as short chain dehydrogenase. For in vivo NADPH analyses a codon-optimized gene for mBFP was introduced into Corynebacterium glutamicum WT and the phosphoglucoisomerase-deficient strain C. glutamicum Δpgi, which accumulates high levels of NADPH. For determination of intracellular NADPH concentrations by mBFP a calibration method with permeabilized cells was developed. By this means an increase of intracellular NADPH concentrations within seconds after the addition of glucose to nutrient-starved cells of both C. glutamicum WT and C. glutamicum Δpgi was observed; as expected the internal NADPH concentration was significantly higher for C. glutamicum Δpgi (0.31 mM) when compared to C. glutamicum WT (0.19 mM). Addition of paraquat to E. coli cells carrying mBFP led as expected to an immediate decrease of intracellular NADPH concentrations, showing the versatile use of mBFP as intracellular sensor.
Keywords: Corynebacterium glutamicum; Escherichia coli; NADPH; biosensor; redox state; short chain dehydrogenase.
Figures


References
-
- Blacker T. S., Mann Z. F., Gale J. E., Ziegler M., Bain A. J., Duchen M. R. (2012). Separation of NADPH and NADH fluorescence emission in live cells using flim. Biophys. J. 102 196a. 10.1016/j.bpj.2011.11.1067 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

