Molecular Evolution and Stress and Phytohormone Responsiveness of SUT Genes in Gossypium hirsutum
- PMID: 30405700
- PMCID: PMC6205988
- DOI: 10.3389/fgene.2018.00494
Molecular Evolution and Stress and Phytohormone Responsiveness of SUT Genes in Gossypium hirsutum
Abstract
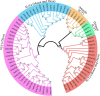
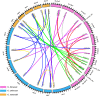
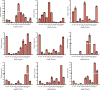
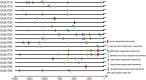
Sucrose transporters (SUTs) play key roles in allocating the translocation of assimilates from source to sink tissues. Although the characteristics and biological roles of SUTs have been intensively investigated in higher plants, this gene family has not been functionally characterized in cotton. In this study, we performed a comprehensive analysis of SUT genes in the tetraploid cotton Gossypium hirsutum. A total of 18 G. hirsutum SUT genes were identified and classified into three groups based on their evolutionary relationships. Up to eight SUT genes in G. hirsutum were placed in the dicot-specific SUT1 group, while four and six SUT genes were, respectively, clustered into SUT4 and SUT2 groups together with members from both dicot and monocot species. The G. hirsutum SUT genes within the same group displayed similar exon/intron characteristics, and homologous genes in G. hirsutum At and Dt subgenomes, G. arboreum, and G. raimondii exhibited one-to-one relationships. Additionally, the duplicated genes in the diploid and polyploid cotton species have evolved through purifying selection, suggesting the strong conservation of SUT loci in these species. Expression analysis in different tissues indicated that SUT genes might play significant roles in cotton fiber elongation. Moreover, analyses of cis-acting regulatory elements in promoter regions and expression profiling under different abiotic stress and exogenous phytohormone treatments implied that SUT genes, especially GhSUT6A/D, might participate in plant responses to diverse abiotic stresses and phytohormones. Our findings provide valuable information for future studies on the evolution and function of SUT genes in cotton.
Keywords: abiotic stress; cotton; expression profile; phylogenetic relationship; phytohormone; sucrose transporter.
Figures


Similar articles
-
Structure, phylogeny, allelic haplotypes and expression of sucrose transporter gene families in Saccharum.BMC Genomics. 2016 Feb 1;17:88. doi: 10.1186/s12864-016-2419-6. BMC Genomics. 2016. PMID: 26830680 Free PMC article.
-
Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses.BMC Genomics. 2018 Aug 9;19(1):599. doi: 10.1186/s12864-018-4985-2. BMC Genomics. 2018. PMID: 30092779 Free PMC article.
-
Genome wide identification, classification and functional characterization of heat shock transcription factors in cultivated and ancestral cottons (Gossypium spp.).Int J Biol Macromol. 2021 Jul 1;182:1507-1527. doi: 10.1016/j.ijbiomac.2021.05.016. Epub 2021 May 7. Int J Biol Macromol. 2021. PMID: 33965497
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Rice SUT and SWEET Transporters.Int J Mol Sci. 2021 Oct 18;22(20):11198. doi: 10.3390/ijms222011198. Int J Mol Sci. 2021. PMID: 34681858 Free PMC article. Review.
Cited by
-
Evolutionary History and Functional Diversification of the JmjC Domain-Containing Histone Demethylase Gene Family in Plants.Plants (Basel). 2022 Apr 12;11(8):1041. doi: 10.3390/plants11081041. Plants (Basel). 2022. PMID: 35448769 Free PMC article.
-
Identification and molecular evolution of the GLX genes in 21 plant species: a focus on the Gossypium hirsutum.BMC Genomics. 2023 Aug 22;24(1):474. doi: 10.1186/s12864-023-09524-w. BMC Genomics. 2023. PMID: 37608304 Free PMC article.
-
Genome-Wide Identification, Plasma Membrane Localization, and Functional Validation of the SUT Gene Family in Yam (Dioscorea cayennensis subsp. rotundata).Int J Mol Sci. 2025 Jun 16;26(12):5756. doi: 10.3390/ijms26125756. Int J Mol Sci. 2025. PMID: 40565219 Free PMC article.
-
Characterization and Stress Response of the JmjC Domain-Containing Histone Demethylase Gene Family in the Allotetraploid Cotton Species Gossypium hirsutum.Plants (Basel). 2020 Nov 20;9(11):1617. doi: 10.3390/plants9111617. Plants (Basel). 2020. PMID: 33233854 Free PMC article.
-
Phylogenetic relationships of sucrose transporters (SUTs) in plants and genome-wide characterization of SUT genes in Orchidaceae reveal roles in floral organ development.PeerJ. 2021 Sep 13;9:e11961. doi: 10.7717/peerj.11961. eCollection 2021. PeerJ. 2021. PMID: 34603845 Free PMC article.
References
LinkOut - more resources
Full Text Sources

