Proteomic Profiling of Burkholderia thailandensis During Host Infection Using Bio-Orthogonal Noncanonical Amino Acid Tagging (BONCAT)
- PMID: 30406044
- PMCID: PMC6206043
- DOI: 10.3389/fcimb.2018.00370
Proteomic Profiling of Burkholderia thailandensis During Host Infection Using Bio-Orthogonal Noncanonical Amino Acid Tagging (BONCAT)
Abstract
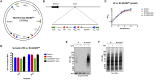
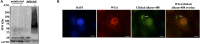
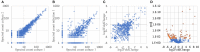
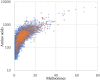
Burkholderia pseudomallei and B. mallei are the causative agents of melioidosis and glanders, respectively, and are often fatal to humans and animals. Owing to the high fatality rate, potential for spread by aerosolization, and the lack of efficacious therapeutics, B. pseudomallei and B. mallei are considered biothreat agents of concern. In this study, we investigate the proteome of Burkholderia thailandensis, a closely related surrogate for the two more virulent Burkholderia species, during infection of host cells, and compare to that of B. thailandensis in culture. Studying the proteome of Burkholderia spp. during infection is expected to reveal molecular mechanisms of intracellular survival and host immune evasion; but proteomic profiling of Burkholderia during host infection is challenging. Proteomic analyses of host-associated bacteria are typically hindered by the overwhelming host protein content recovered from infected cultures. To address this problem, we have applied bio-orthogonal noncanonical amino acid tagging (BONCAT) to B. thailandensis, enabling the enrichment of newly expressed bacterial proteins from virtually any growth condition, including host cell infection. In this study, we show that B. thailandensis proteins were selectively labeled and efficiently enriched from infected host cells using BONCAT. We also demonstrate that this method can be used to label bacteria in situ by fluorescent tagging. Finally, we present a global proteomic profile of B. thailandensis as it infects host cells and a list of proteins that are differentially regulated in infection conditions as compared to bacterial monoculture. Among the identified proteins are quorum sensing regulated genes as well as homologs to previously identified virulence factors. This method provides a powerful tool to study the molecular processes during Burkholderia infection, a much-needed addition to the Burkholderia molecular toolbox.
Keywords: BONCAT; Burkholderia; host infection; intracellular pathogen; orthogonal amino acid labeling; protein enrichment; proteome profiling; quorum sensing (QS).
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

