DNA Sliding Clamps as Therapeutic Targets
- PMID: 30406112
- PMCID: PMC6204406
- DOI: 10.3389/fmolb.2018.00087
DNA Sliding Clamps as Therapeutic Targets
Abstract
Chromosomal DNA replication is achieved by an assembly of multi-protein complexes at the replication fork. DNA sliding clamps play an important role in this assembly and are essential for cell viability. Inhibitors of bacterial (β-clamp) and eukaryal DNA clamps, proliferating cell nuclear antigen (PCNA), have been explored for use as antibacterial and anti-cancer drugs, respectively. Inhibitors for bacterial β-clamps include modified peptides, small molecule inhibitors, natural products, and modified non-steroidal anti-inflammatory drugs. Targeting eukaryotic PCNA sliding clamp in its role in replication can be complicated by undesired effects on healthy cells. Some success has been seen in the design of peptide inhibitors, however, other research has focused on targeting PCNA molecules that are modified in diseased states. These inhibitors that are targeted to PCNA involved in DNA repair can sensitize cancer cells to existing anti-cancer therapeutics, and a DNA aptamer has also been shown to inhibit PCNA. In this review, studies in the use of both bacterial and eukaryotic sliding clamps as therapeutic targets are summarized.
Keywords: DNA clamp; DNA sliding clamp; PCNA; proliferating cell nuclear antigen; therapeutic; β-clamp.
Figures
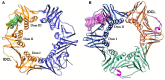
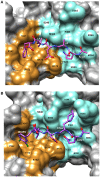
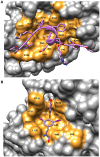
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

