Zic4-Lineage Cells Increase Their Contribution to Visual Thalamic Nuclei during Murine Embryogenesis If They Are Homozygous or Heterozygous for Loss of Pax6 Function
- PMID: 30406191
- PMCID: PMC6220585
- DOI: 10.1523/ENEURO.0367-18.2018
Zic4-Lineage Cells Increase Their Contribution to Visual Thalamic Nuclei during Murine Embryogenesis If They Are Homozygous or Heterozygous for Loss of Pax6 Function
Abstract
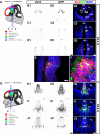
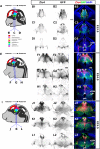
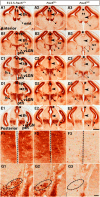
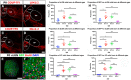
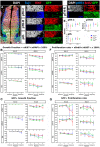
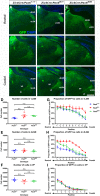
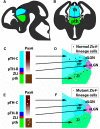
Our aim was to study the mechanisms that contribute to the development of discrete thalamic nuclei during mouse embryogenesis (both sexes included). We characterized the expression of the transcription factor coding gene Zic4 and the distribution of cells that expressed Zic4 in their lineage. We used genetic fate mapping to show that Zic4-lineage cells mainly contribute to a subset of thalamic nuclei, in particular the lateral geniculate nuclei (LGNs), which are crucial components of the visual pathway. We observed that almost all Zic4-lineage diencephalic progenitors express the transcription factor Pax6 at variable location-dependent levels. We used conditional mutagenesis to delete either one or both copies of Pax6 from Zic4-lineage cells. We found that Zic4-lineage cells carrying either homozygous or heterozygous loss of Pax6 contributed in abnormally high numbers to one or both of the main lateral geniculate nuclei (LGNs). This could not be attributed to a change in cell production and was likely due to altered sorting of thalamic cells. Our results indicate that positional information encoded by the levels of Pax6 in diencephalic progenitors is an important determinant of the eventual locations of their daughter cells.
Keywords: Pax6; Zic4; lateral geniculate nucleus; prethalamus; thalamus.
Figures





Similar articles
-
Pax6 regulates the formation of the habenular nuclei by controlling the temporospatial expression of Shh in the diencephalon in vertebrates.BMC Biol. 2014 Feb 14;12:13. doi: 10.1186/1741-7007-12-13. BMC Biol. 2014. PMID: 24528677 Free PMC article.
-
Differential gene expression in the developing lateral geniculate nucleus and medial geniculate nucleus reveals novel roles for Zic4 and Foxp2 in visual and auditory pathway development.J Neurosci. 2009 Oct 28;29(43):13672-83. doi: 10.1523/JNEUROSCI.2127-09.2009. J Neurosci. 2009. PMID: 19864579 Free PMC article.
-
Pax6 is required at the telencephalic pallial-subpallial boundary for the generation of neuronal diversity in the postnatal limbic system.J Neurosci. 2011 Apr 6;31(14):5313-24. doi: 10.1523/JNEUROSCI.3867-10.2011. J Neurosci. 2011. PMID: 21471366 Free PMC article.
-
Regulation of eye formation by the Rx and pax6 homeobox genes.Cell Mol Life Sci. 2000 Feb;57(2):186-94. doi: 10.1007/PL00000683. Cell Mol Life Sci. 2000. PMID: 10766016 Free PMC article. Review.
-
Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens.Int J Dev Biol. 2004;48(8-9):829-44. doi: 10.1387/ijdb.041866ac. Int J Dev Biol. 2004. PMID: 15558475 Free PMC article. Review.
Cited by
-
The role of the diencephalon in the guidance of thalamocortical axons in mice.Development. 2020 Jun 26;147(12):dev184523. doi: 10.1242/dev.184523. Development. 2020. PMID: 32541009 Free PMC article.
-
Development of the early fetal human thalamus: from a protomap to emergent thalamic nuclei.Front Neuroanat. 2025 Feb 7;19:1530236. doi: 10.3389/fnana.2025.1530236. eCollection 2025. Front Neuroanat. 2025. PMID: 39990522 Free PMC article.
References
-
- Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman VM, Mikoshiba K (1996a) The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J Biol Chem 271:1043–1047. - PubMed
-
- Aruga J, Yozu A, Hayashizaki Y, Okazaki Y, Chapman VM, Mikoshiba K (1996b) Identification and characterization of Zic4, a new member of the mouse Zic gene family. Gene 172:291–294. - PubMed
-
- Bamiou DE, Free SL, Sisodiya SM, Chong WK, Musiek F, Williamson KA, van Heyningen V, Moore AT, Gadian D, Luxon LM (2007) Auditory interhemispheric transfer deficits, hearing difficulties, and brain magnetic resonance imaging abnormalities in children with congenital aniridia due to PAX6 mutations. Arch Pediatr Adolesc Med 161:463–469. 10.1001/archpedi.161.5.463 - DOI - PubMed
-
- Caballero IM, Manuel MN, Molinek M, Quintana-Urzainqui I, Mi D, Shimogori T, Price DJ (2014) Cell-autonomous repression of Shh by transcription factor Pax6 regulates diencephalic patterning by controlling the central diencephalic organizer. Cell Rep 8:1405–1418. 10.1016/j.celrep.2014.07.051 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
