Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB
- PMID: 30407520
- PMCID: PMC6323967
- DOI: 10.1093/nar/gky1084
Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB
Abstract
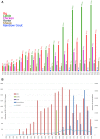
Successful development of biological databases requires accommodation of the burgeoning amounts of data from high-throughput genomics pipelines. As the volume of curated data in Animal QTLdb (https://www.animalgenome.org/QTLdb) increases exponentially, the resulting challenges must be met with rapid infrastructure development to effectively accommodate abundant data curation and make metadata analysis more powerful. The development of Animal QTLdb and CorrDB for the past 15 years has provided valuable tools for researchers to utilize a wealth of phenotype/genotype data to study the genetic architecture of livestock traits. We have focused our efforts on data curation, improved data quality maintenance, new tool developments, and database co-developments, in order to provide convenient platforms for users to query and analyze data. The database currently has 158 499 QTL/associations, 10 482 correlations and 1977 heritability data as a result of an average 32% data increase per year. In addition, we have made >14 functional improvements or new tool implementations since our last report. Our ultimate goals of database development are to provide infrastructure for data collection, curation, and annotation, and more importantly, to support innovated data structure for new types of data mining, data reanalysis, and networked genetic analysis that lead to the generation of new knowledge.
Figures





References
-
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004; 432:695–716. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

