Update of the FANTOM web resource: expansion to provide additional transcriptome atlases
- PMID: 30407557
- PMCID: PMC6323950
- DOI: 10.1093/nar/gky1099
Update of the FANTOM web resource: expansion to provide additional transcriptome atlases
Abstract
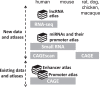
The FANTOM web resource (http://fantom.gsc.riken.jp/) was developed to provide easy access to the data produced by the FANTOM project. It contains the most complete and comprehensive sets of actively transcribed enhancers and promoters in the human and mouse genomes. We determined the transcription activities of these regulatory elements by CAGE (Cap Analysis of Gene Expression) for both steady and dynamic cellular states in all major and some rare cell types, consecutive stages of differentiation and responses to stimuli. We have expanded the resource by employing different assays, such as RNA-seq, short RNA-seq and a paired-end protocol for CAGE (CAGEscan), to provide new angles to study the transcriptome. That yielded additional atlases of long noncoding RNAs, miRNAs and their promoters. We have also expanded the CAGE analysis to cover rat, dog, chicken, and macaque species for a limited number of cell types. The CAGE data obtained from human and mouse were reprocessed to make them available on the latest genome assemblies. Here, we report the recent updates of both data and interfaces in the FANTOM web resource.
Figures



Similar articles
-
FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs.Nucleic Acids Res. 2021 Jan 8;49(D1):D892-D898. doi: 10.1093/nar/gkaa1054. Nucleic Acids Res. 2021. PMID: 33211864 Free PMC article.
-
Update of the FANTOM web resource: enhancement for studying noncoding genomes.Nucleic Acids Res. 2025 Jan 6;53(D1):D419-D424. doi: 10.1093/nar/gkae1047. Nucleic Acids Res. 2025. PMID: 39592010 Free PMC article.
-
Update of the FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation.Nucleic Acids Res. 2011 Jan;39(Database issue):D856-60. doi: 10.1093/nar/gkq1112. Epub 2010 Nov 12. Nucleic Acids Res. 2011. PMID: 21075797 Free PMC article.
-
Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine.Curr Top Microbiol Immunol. 2016;394:237-58. doi: 10.1007/82_2015_464. Curr Top Microbiol Immunol. 2016. PMID: 26318140 Review.
-
lncRNAs and microRNAs with a role in cancer development.Biochim Biophys Acta. 2016 Jan;1859(1):169-76. doi: 10.1016/j.bbagrm.2015.06.015. Epub 2015 Jul 4. Biochim Biophys Acta. 2016. PMID: 26149773 Review.
Cited by
-
Translation of genome-wide association study: from genomic signals to biological insights.Front Genet. 2024 Oct 3;15:1375481. doi: 10.3389/fgene.2024.1375481. eCollection 2024. Front Genet. 2024. PMID: 39421299 Free PMC article. Review.
-
Potential Tumor Suppressor Role of Polo-like Kinase 5 in Cancer.Cancers (Basel). 2023 Nov 17;15(22):5457. doi: 10.3390/cancers15225457. Cancers (Basel). 2023. PMID: 38001717 Free PMC article.
-
CALML5 is a novel diagnostic marker for differentiating thymic squamous cell carcinoma from type B3 thymoma.Thorac Cancer. 2023 Apr;14(12):1089-1097. doi: 10.1111/1759-7714.14853. Epub 2023 Mar 16. Thorac Cancer. 2023. PMID: 36924358 Free PMC article.
-
Defining the fine structure of promoter activity on a genome-wide scale with CISSECTOR.Nucleic Acids Res. 2023 Jun 23;51(11):5499-5511. doi: 10.1093/nar/gkad232. Nucleic Acids Res. 2023. PMID: 37013986 Free PMC article.
-
Functional non-coding RNAs in vascular diseases.FEBS J. 2021 Nov;288(22):6315-6330. doi: 10.1111/febs.15678. Epub 2021 Jan 7. FEBS J. 2021. PMID: 33340430 Free PMC article. Review.
References
-
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C. et al. . Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006; 38:626–635. - PubMed
-
- FANTOM Consortium Suzuki H., Forrest A.R., van Nimwegen E., Daub C.O., Balwierz P.J., Irvine K.M., Lassmann T., Ravasi T., Hasegawa Y. et al. . The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009; 41:553–562. - PMC - PubMed
-
- Kawai J., Shinagawa A., Shibata K., Yoshino M., Itoh M., Ishii Y., Arakawa T., Hara A., Fukunishi Y., Konno H. et al. . Functional annotation of a full-length mouse cDNA collection. Nature. 2001; 409:685–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

