Resequencing of cv CRI-12 family reveals haplotype block inheritance and recombination of agronomically important genes in artificial selection
- PMID: 30407717
- PMCID: PMC6587942
- DOI: 10.1111/pbi.13030
Resequencing of cv CRI-12 family reveals haplotype block inheritance and recombination of agronomically important genes in artificial selection
Abstract
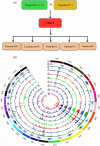
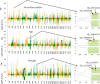
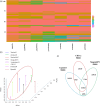
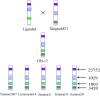
Although efforts have been taken to exploit diversity for yield and quality improvements, limited progress on using beneficial alleles in domesticated and undomesticated cotton varieties is limited. Given the complexity and limited amount of genomic information since the completion of four cotton genomes, characterizing significant variations and haplotype block inheritance under artificial selection has been challenging. Here we sequenced Gossypium hirsutum L. cv CRI-12 (the cotton variety with the largest acreage in China), its parental cultivars, and progeny cultivars, which were bred by the different institutes in China. In total, 3.3 million SNPs were identified and 118, 126 and 176 genes were remarkably correlated with Verticillium wilt, salinity and drought tolerance in CRI-12, respectively. Transcriptome-wide analyses of gene expression, and functional annotations, have provided support for the identification of genes tied to these tolerances. We totally discovered 58 116 haplotype blocks, among which 23 752 may be inherited and 1029 may be recombined under artificial selection. This survey of genetic diversity identified loci that may have been subject to artificial selection and documented the haplotype block inheritance and recombination, shedding light on the genetic mechanism of artificial selection and guiding breeding efforts for the genetic improvement of cotton.
Keywords: CRI-12 family; SNPs; artificial selection; haplotype block; inheritance and recombination.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures
Similar articles
-
QTL analysis for yield and fibre quality traits using three sets of introgression lines developed from three Gossypium hirsutum race stocks.Mol Genet Genomics. 2019 Jun;294(3):789-810. doi: 10.1007/s00438-019-01548-w. Epub 2019 Mar 18. Mol Genet Genomics. 2019. PMID: 30887144
-
Genetic and transcriptomic dissection of the fiber length trait from a cotton (Gossypium hirsutum L.) MAGIC population.BMC Genomics. 2019 Feb 6;20(1):112. doi: 10.1186/s12864-019-5427-5. BMC Genomics. 2019. PMID: 30727946 Free PMC article.
-
Detection of candidate genes and development of KASP markers for Verticillium wilt resistance by combining genome-wide association study, QTL-seq and transcriptome sequencing in cotton.Theor Appl Genet. 2021 Apr;134(4):1063-1081. doi: 10.1007/s00122-020-03752-4. Epub 2021 Jan 12. Theor Appl Genet. 2021. PMID: 33438060
-
Two genomic regions associated with fiber quality traits in Chinese upland cotton under apparent breeding selection.Sci Rep. 2016 Dec 7;6:38496. doi: 10.1038/srep38496. Sci Rep. 2016. PMID: 27924947 Free PMC article.
-
Deepening genomic sequences of 1081 Gossypium hirsutum accessions reveals novel SNPs and haplotypes relevant for practical breeding utility.Genomics. 2024 Jul;116(4):110848. doi: 10.1016/j.ygeno.2024.110848. Epub 2024 Apr 24. Genomics. 2024. PMID: 38663523
Cited by
-
Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum.Genes (Basel). 2019 Mar 18;10(3):226. doi: 10.3390/genes10030226. Genes (Basel). 2019. PMID: 30889904 Free PMC article.
-
Breeding crops by design for future agriculture.J Zhejiang Univ Sci B. 2020 Jun;21(6):423-425. doi: 10.1631/jzus.B2010001. J Zhejiang Univ Sci B. 2020. PMID: 32478489 Free PMC article.
-
EST-SNP Study of Olea europaea L. Uncovers Functional Polymorphisms between Cultivated and Wild Olives.Genes (Basel). 2020 Aug 10;11(8):916. doi: 10.3390/genes11080916. Genes (Basel). 2020. PMID: 32785094 Free PMC article.
-
Dissection of the Pearl of Csaba pedigree identifies key genomic segments related to early ripening in grape.Plant Physiol. 2023 Feb 12;191(2):1153-1166. doi: 10.1093/plphys/kiac539. Plant Physiol. 2023. PMID: 36440478 Free PMC article.
-
A high-quality assembled genome and its comparative analysis decode the adaptive molecular mechanism of the number one Chinese cotton variety CRI-12.Gigascience. 2022 Apr 1;11:giac019. doi: 10.1093/gigascience/giac019. Gigascience. 2022. PMID: 35365835 Free PMC article.
References
-
- Barrett, J.C. , Fry, B. , Maller, J. and Daly, M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

