AFLP-AFLP in silico-NGS approach reveals polymorphisms in repetitive elements in the malignant genome
- PMID: 30408048
- PMCID: PMC6224067
- DOI: 10.1371/journal.pone.0206620
AFLP-AFLP in silico-NGS approach reveals polymorphisms in repetitive elements in the malignant genome
Abstract
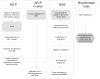
The increasing interest in exploring the human genome and identifying genetic risk factors contributing to the susceptibility to and outcome of diseases has supported the rapid development of genome-wide techniques. However, the large amount of obtained data requires extensive bioinformatics analysis. In this work, we established an approach combining amplified fragment length polymorphism (AFLP), AFLP in silico and next generation sequencing (NGS) methods to map the malignant genome of patients with chronic myeloid leukemia. We compared the unique DNA fingerprints of patients generated by the AFLP technique approach with those of healthy donors to identify AFLP markers associated with the disease and/or the response to treatment with imatinib, a tyrosine kinase inhibitor. Among the statistically significant AFLP markers selected for NGS analysis and virtual fingerprinting, we identified the sequences of three fragments in the region of DNA repeat element OldhAT1, LINE L1M7, LTR MER90, and satellite ALR/Alpha among repetitive elements, which may indicate a role of these non-coding repetitive sequences in hematological malignancy. SNPs leading to the presence/absence of these fragments were confirmed by Sanger sequencing. When evaluating the results of AFLP analysis for some fragments, we faced the frequently discussed size homoplasy, resulting in co-migration of non-identical AFLP fragments that may originate from an insertion/deletion, SNP, somatic mutation anywhere in the genome, or combination thereof. The AFLP-AFLP in silico-NGS procedure represents a smart alternative to microarrays and relatively expensive and bioinformatically challenging whole-genome sequencing to detect the association of variable regions of the human genome with diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Next-generation sequencing for sensitive detection of BCR-ABL1 mutations relevant to tyrosine kinase inhibitor choice in imatinib-resistant patients.Oncotarget. 2016 Apr 19;7(16):21982-90. doi: 10.18632/oncotarget.8010. Oncotarget. 2016. PMID: 26980736 Free PMC article.
-
In silico fingerprinting (ISIF): a user-friendly in silico AFLP program.Methods Mol Biol. 2012;888:55-64. doi: 10.1007/978-1-61779-870-2_4. Methods Mol Biol. 2012. PMID: 22665275
-
Two methods to easily obtain nucleotide sequences from AFLP loci of interest.Methods Mol Biol. 2012;888:91-108. doi: 10.1007/978-1-61779-870-2_6. Methods Mol Biol. 2012. PMID: 22665277
-
[MET/ERK and MET/JNK Pathway Activation Is Involved in BCR-ABL Inhibitor-resistance in Chronic Myeloid Leukemia].Yakugaku Zasshi. 2018;138(12):1461-1466. doi: 10.1248/yakushi.18-00142. Yakugaku Zasshi. 2018. PMID: 30504658 Review. Japanese.
-
Amplified fragment length polymorphism: an adept technique for genome mapping, genetic differentiation, and intraspecific variation in protozoan parasites.Parasitol Res. 2013 Feb;112(2):457-66. doi: 10.1007/s00436-012-3238-6. Epub 2012 Dec 20. Parasitol Res. 2013. PMID: 23254590 Review.
References
-
- de Brevern AG, Meyniel J, Fairhead C, Neuvéglise C, Malpertuy A. Trends in IT Innovation to Build a Next Generation Bioinformatics Solution to Manage and Analyse Biological Big Data Produced by NGS Technologies. BioMed research international. 2015;2015: 904541 10.1155/2015/904541 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

