Measuring radiation-induced DNA damage in Cryptococcus neoformans and Saccharomyces cerevisiae using long range quantitative PCR
- PMID: 30408089
- PMCID: PMC6224075
- DOI: 10.1371/journal.pone.0207071
Measuring radiation-induced DNA damage in Cryptococcus neoformans and Saccharomyces cerevisiae using long range quantitative PCR
Abstract
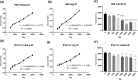
DNA damage has been considered to be the universal critical lesion in cells after exposure to ionizing radiation. Measuring radiation-induced DNA damage is important to understand the mechanisms of radiation-induced toxicity and monitor DNA damage repairs. Currently the most widely used methods to measure DNA damage are pulsed-field gel electrophoresis (PFGF) and single-cell gel electrophoresis (also known as the comet assay), both of which are technically challenging and time consuming. Long range quantitative polymerase chain reaction (LR-QPCR) has been used successfully to measure nuclear and mitochondrial DNA damage in mammalian and several model organism cells. The principle of this assay is that DNA lesions will slow down or block the progression of DNA polymerase. Therefore, the amplification efficiency of DNA with fewer lesions will be higher than DNA with more lesions under the same reaction condition. Here, we developed the LR-QPCR assay primers and reaction conditions to quantify DNA damage in Cryptococcus neoformans (C. neoformans) and Saccharomyces cerevisiae (S. cerevisiae) after gamma ray exposure. Under these conditions, long DNA targets of C. neoformans H99 and S. cerevisiae BY4741 (17.6 and 16.4 kb for nuclear DNA and 15.3 and 14.6 kb for mitochondrial DNA) were quantitatively amplified using extracted DNA templates, respectively. Two short mitochondrial DNA targets of these two species (207 bp and 154 bp) were also quantitatively amplified and used to monitor the number of mitochondria. Using the LR-QPCR method, we showed that the frequency of radiation-induced mitochondrial and nuclear DNA lesions had a significant linear correlation with the radiation doses (from 500 Gy to 3000 Gy) in both species. Furthermore, the faster disappearance of DNA damage detected in C. neoformans H99S strain compared to H99 strain may help to explain the different radiation sensitivity of these two strains. In summary, we developed a simple, sensitive method to measure radiation-induced DNA damage, which can greatly facilitate the study of radiation-induced toxicity and can be widely used as a dosimetry in radiation-induced cell damage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Unraveling Fungal Radiation Resistance Regulatory Networks through the Genome-Wide Transcriptome and Genetic Analyses of Cryptococcus neoformans.mBio. 2016 Nov 29;7(6):e01483-16. doi: 10.1128/mBio.01483-16. mBio. 2016. PMID: 27899501 Free PMC article.
-
Effects of radiation type and delivery mode on a radioresistant eukaryote Cryptococcus neoformans.Nucl Med Biol. 2015 Jun;42(6):515-23. doi: 10.1016/j.nucmedbio.2015.02.006. Epub 2015 Mar 11. Nucl Med Biol. 2015. PMID: 25800676 Free PMC article.
-
Identification and properties of plasma membrane azole efflux pumps from the pathogenic fungi Cryptococcus gattii and Cryptococcus neoformans.J Antimicrob Chemother. 2015 May;70(5):1396-407. doi: 10.1093/jac/dku554. Epub 2015 Jan 27. J Antimicrob Chemother. 2015. PMID: 25630649 Free PMC article.
-
[Cell cycle control and CDC28/Cdc2 homologue and related gene cloning of Cryptococcus neoformans].Nihon Ishinkin Gakkai Zasshi. 2006;47(4):257-62. doi: 10.3314/jjmm.47.257. Nihon Ishinkin Gakkai Zasshi. 2006. PMID: 17086156 Review. Japanese.
-
The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number.Methods. 2010 Aug;51(4):444-51. doi: 10.1016/j.ymeth.2010.01.033. Epub 2010 Feb 1. Methods. 2010. PMID: 20123023 Free PMC article. Review.
Cited by
-
Proanthocyanidins Ameliorated Deficits of Lipid Metabolism in Type 2 Diabetes Mellitus Via Inhibiting Adipogenesis and Improving Mitochondrial Function.Int J Mol Sci. 2020 Mar 16;21(6):2029. doi: 10.3390/ijms21062029. Int J Mol Sci. 2020. PMID: 32188147 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

