A Mass-Spectrometry-Based Modelling Workflow for Accurate Prediction of IgG Antibody Conformations in the Gas Phase
- PMID: 30408305
- PMCID: PMC6392142
- DOI: 10.1002/anie.201812018
A Mass-Spectrometry-Based Modelling Workflow for Accurate Prediction of IgG Antibody Conformations in the Gas Phase
Abstract
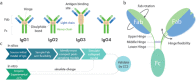
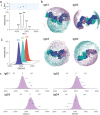
Immunoglobulins are biomolecules involved in defence against foreign substances. Flexibility is key to their functional properties in relation to antigen binding and receptor interactions. We have developed an integrative strategy combining ion mobility mass spectrometry (IM-MS) with molecular modelling to study the conformational dynamics of human IgG antibodies. Predictive models of all four human IgG subclasses were assembled and their dynamics sampled in the transition from extended to collapsed state during IM-MS. Our data imply that this collapse of IgG antibodies is related to their intrinsic structural features, including Fab arm flexibility, collapse towards the Fc region, and the length of their hinge regions. The workflow presented here provides an accurate structural representation in good agreement with the observed collision cross section for these flexible IgG molecules. These results have implications for studying other nonglobular flexible proteins.
Keywords: conformation analysis; immunoglobulin; ion mobility; mass spectrometry; molecular dynamics.
© 2018 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Native Mass Spectrometry, Ion Mobility, and Collision-Induced Unfolding for Conformational Characterization of IgG4 Monoclonal Antibodies.Anal Chem. 2018 Aug 7;90(15):8865-8872. doi: 10.1021/acs.analchem.8b00912. Epub 2018 Jul 16. Anal Chem. 2018. PMID: 29956914
-
Molecular Insights into the Thermal Stability of mAbs with Variable-Temperature Ion-Mobility Mass Spectrometry.Chembiochem. 2016 Jan 1;17(1):46-51. doi: 10.1002/cbic.201500574. Epub 2015 Dec 3. Chembiochem. 2016. PMID: 26534882
-
Investigating the interaction between the neonatal Fc receptor and monoclonal antibody variants by hydrogen/deuterium exchange mass spectrometry.Mol Cell Proteomics. 2015 Jan;14(1):148-61. doi: 10.1074/mcp.M114.042044. Epub 2014 Nov 6. Mol Cell Proteomics. 2015. PMID: 25378534 Free PMC article.
-
How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase.Analyst. 2011 Jan 7;136(1):20-8. doi: 10.1039/c0an00373e. Epub 2010 Aug 31. Analyst. 2011. PMID: 20820495 Review.
-
[Applications of ion mobility-mass spectrometry in the chemical analysis in traditional Chinese medicines].Se Pu. 2022 Sep;40(9):782-787. doi: 10.3724/SP.J.1123.2022.01028. Se Pu. 2022. PMID: 36156624 Free PMC article. Review. Chinese.
Cited by
-
Are the Gas-Phase Structures of Molecular Elephants Enduring or Ephemeral? Results from Time-Dependent, Tandem Ion Mobility.Anal Chem. 2023 Jun 27;95(25):9589-9597. doi: 10.1021/acs.analchem.3c01222. Epub 2023 Jun 9. Anal Chem. 2023. PMID: 37294019 Free PMC article.
-
High-Throughput Multi-attribute Analysis of Antibody-Drug Conjugates Enabled by Trapped Ion Mobility Spectrometry and Top-Down Mass Spectrometry.Anal Chem. 2021 Jul 27;93(29):10013-10021. doi: 10.1021/acs.analchem.1c00150. Epub 2021 Jul 14. Anal Chem. 2021. PMID: 34258999 Free PMC article.
-
Rapid structural discrimination of IgG antibodies by multicharge-state collision-induced unfolding.RSC Adv. 2021 Nov 12;11(58):36502-36510. doi: 10.1039/d1ra06486j. eCollection 2021 Nov 10. RSC Adv. 2021. PMID: 35494361 Free PMC article.
-
Defining the mobility range of a hinge-type connection using molecular dynamics and metadynamics.PLoS One. 2020 Apr 13;15(4):e0230962. doi: 10.1371/journal.pone.0230962. eCollection 2020. PLoS One. 2020. PMID: 32282813 Free PMC article.
-
Surface-induced Dissociation Mass Spectrometry as a Structural Biology Tool.Chem Rev. 2022 Apr 27;122(8):7442-7487. doi: 10.1021/acs.chemrev.1c00309. Epub 2021 Nov 2. Chem Rev. 2022. PMID: 34726898 Free PMC article. Review.
References
-
- Irani V., Guy A. J., Andrew D., Beeson J. G., Ramsland P. A., Richards J. S., Mol. Immunol. 2015, 67, 171–182. - PubMed
-
- Jakobovits A., Amado R. G., Yang X., Roskos L., Schwab G., Nat. Biotechnol. 2007, 25, 1134–1143. - PubMed
-
- Murphy K., Weaver C., Mowat A., Berg L., Chaplin D. D., Janeway's immunobiology, 9th ed., Garland Science/Taylor & Francis Group, LLC, New York, NY, 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

