Investigation on the processing and improving the cleavage efficiency of furin cleavage sites in Pichia pastoris
- PMID: 30409181
- PMCID: PMC6223083
- DOI: 10.1186/s12934-018-1020-x
Investigation on the processing and improving the cleavage efficiency of furin cleavage sites in Pichia pastoris
Abstract
Background: Proprotein convertase furin is responsible for the processing of a wide variety of precursors consisted of signal peptide, propeptide and mature peptide in mammal. Many precursors processed by furin have important physiological functions and can be recombinantly expressed in Pichia pastoris expression system for research, pharmaceutical and vaccine applications. However, it is not clear whether the furin cleavage sites between the propeptide and mature peptide can be properly processed in P. pastoris, bringing uncertainty for proper expression of the coding DNA sequences of furin precursors containing the propeptides and mature peptides.
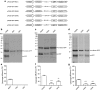
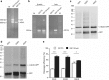
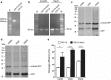
Results: In this study, we evaluated the ability of P. pastoris to process furin cleavage sites and how to improve the cleavage efficiencies of furin cleavage sites in P. pastoris. The results showed that P. pastoris can process furin cleavage sites but the cleavage efficiencies are not high. Arg residue at position P1 or P4 in furin cleavage sites significantly affect cleavage efficiency in P. pastoris. Kex2 protease, but not YPS1, in P. pastoris is responsible for processing furin cleavage sites. Heterologous expression of furin or overexpression of Kex2 in P. pastoris effectively increased cleavage efficiencies of furin cleavage sites.
Conclusions: Our investigation on the processing of furin cleavage sites provides important information for recombinant expression of furin precursors in P. pastoris. Furin or Kex2 overexpressing strains may be good choices for expressing precursors processed by furin in P. pastoris.
Keywords: Furin; Furin cleavage site; Kex2; Pichia pastoris; Proprotein convertase; YPS1.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

