Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973
- PMID: 30409802
- PMCID: PMC6294925
- DOI: 10.1073/pnas.1814912115
Comparative genomics reveals the molecular determinants of rapid growth of the cyanobacterium Synechococcus elongatus UTEX 2973
Abstract
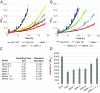
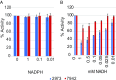
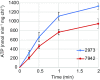
Cyanobacteria are emerging as attractive organisms for sustainable bioproduction. We previously described Synechococcus elongatus UTEX 2973 as the fastest growing cyanobacterium known. Synechococcus 2973 exhibits high light tolerance and an increased photosynthetic rate and produces biomass at three times the rate of its close relative, the model strain Synechococcus elongatus 7942. The two strains differ at 55 genetic loci, and some of these loci must contain the genetic determinants of rapid photoautotrophic growth and improved photosynthetic rate. Using CRISPR/Cpf1, we performed a comprehensive mutational analysis of Synechococcus 2973 and identified three specific genes, atpA, ppnK, and rpaA, with SNPs that confer rapid growth. The fast-growth-associated allele of each gene was then used to replace the wild-type alleles in Synechococcus 7942. Upon incorporation, each allele successively increased the growth rate of Synechococcus 7942; remarkably, inclusion of all three alleles drastically reduced the doubling time from 6.8 to 2.3 hours. Further analysis revealed that our engineering effort doubled the photosynthetic productivity of Synechococcus 7942. We also determined that the fast-growth-associated allele of atpA yielded an ATP synthase with higher specific activity, while that of ppnK encoded a NAD+ kinase with significantly improved kinetics. The rpaA SNPs cause broad changes in the transcriptional profile, as this gene is the master output regulator of the circadian clock. This pioneering study has revealed the molecular basis for rapid growth, demonstrating that limited genetic changes can dramatically improve the growth rate of a microbe by as much as threefold.
Keywords: Synechococcus; cyanobacteria; growth; photosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
Navigating the fitness landscape using multiallele genome editing.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):12547-12549. doi: 10.1073/pnas.1818285115. Epub 2018 Nov 21. Proc Natl Acad Sci U S A. 2018. PMID: 30463944 Free PMC article. No abstract available.
-
Reply to Zhou and Li: Plasticity of the genomic haplotype of Synechococcus elongatus leads to rapid strain adaptation under laboratory conditions.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):3946-3947. doi: 10.1073/pnas.1900792116. Epub 2019 Feb 12. Proc Natl Acad Sci U S A. 2019. PMID: 30755539 Free PMC article. No abstract available.
-
SNPs deciding the rapid growth of cyanobacteria are alterable.Proc Natl Acad Sci U S A. 2019 Mar 5;116(10):3945. doi: 10.1073/pnas.1900210116. Epub 2019 Feb 12. Proc Natl Acad Sci U S A. 2019. PMID: 30755540 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

