Characterization and Genomic Analyses of Pseudomonas aeruginosa Podovirus TC6: Establishment of Genus Pa11virus
- PMID: 30410478
- PMCID: PMC6209634
- DOI: 10.3389/fmicb.2018.02561
Characterization and Genomic Analyses of Pseudomonas aeruginosa Podovirus TC6: Establishment of Genus Pa11virus
Abstract
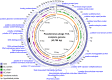
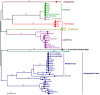
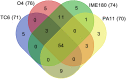
Phages have attracted a renewed interest as alternative to chemical antibiotics. Although the number of phages is 10-fold higher than that of bacteria, the number of genomically characterized phages is far less than that of bacteria. In this study, phage TC6, a novel lytic virus of Pseudomonas aeruginosa, was isolated and characterized. TC6 consists of an icosahedral head with a diameter of approximately 54 nm and a short tail with a length of about 17 nm, which are characteristics of the family Podoviridae. TC6 can lyse 86 out of 233 clinically isolated P. aeruginosa strains, thus showing application potentials for phage therapy. The linear double-stranded genomic DNA of TC6 consisted of 49796 base pairs and was predicted to contain 71 protein-coding genes. A total of 11 TC6 structural proteins were identified by mass spectrometry. Comparative analysis revealed that the P. aeruginosa phages TC6, O4, PA11, and IME180 shared high similarity at DNA sequence and proteome levels, among which PA11 was the first phage discovered and published. Meanwhile, these phages contain 54 core genes and have very close phylogenetic relationships, which distinguish them from other known phage genera. We therefore proposed that these four phages can be classified as Pa11virus, comprising a new phage genus of Podoviridae that infects Pseudomonas spp. The results of this work promoted our understanding of phage biology, classification, and diversity.
Keywords: Pa11virus; Pseudomonas aeruginosa; comparative genomic analysis; phage TC6; podovirus; structural protein.
Figures





Similar articles
-
Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages.PLoS One. 2013 May 13;8(5):e62933. doi: 10.1371/journal.pone.0062933. Print 2013. PLoS One. 2013. PMID: 23675441 Free PMC article.
-
Characterization and Comparative Genomic Analyses of Pseudomonas aeruginosa Phage PaoP5: New Members Assigned to PAK_P1-like Viruses.Sci Rep. 2016 Sep 23;6:34067. doi: 10.1038/srep34067. Sci Rep. 2016. PMID: 27659070 Free PMC article.
-
Characterization of a novel lytic podovirus O4 of Pseudomonas aeruginosa.Arch Virol. 2018 Sep;163(9):2377-2383. doi: 10.1007/s00705-018-3866-y. Epub 2018 May 10. Arch Virol. 2018. PMID: 29749589
-
Characterization and genomic study of "phiKMV-Like" phage PAXYB1 infecting Pseudomonas aeruginosa.Sci Rep. 2017 Oct 12;7(1):13068. doi: 10.1038/s41598-017-13363-7. Sci Rep. 2017. PMID: 29026171 Free PMC article.
-
Bacteriophages of Pseudomonas aeruginosa: long-term prospects for use in phage therapy.Adv Virus Res. 2014;88:227-78. doi: 10.1016/B978-0-12-800098-4.00005-2. Adv Virus Res. 2014. PMID: 24373314 Review.
Cited by
-
Characterization and genome analysis of Pseudomonas aeruginosa phage vB_PaeP_Lx18 and the antibacterial activity of its lysozyme.Arch Virol. 2022 Sep;167(9):1805-1817. doi: 10.1007/s00705-022-05472-0. Epub 2022 Jun 18. Arch Virol. 2022. PMID: 35716268
-
LysSYL: a broad-spectrum phage endolysin targeting Staphylococcus species and eradicating S. aureus biofilms.Microb Cell Fact. 2024 Mar 25;23(1):89. doi: 10.1186/s12934-024-02359-4. Microb Cell Fact. 2024. PMID: 38528536 Free PMC article.
-
Characterization of a Lytic Bacteriophage against Pseudomonas syringae pv. actinidiae and Its Endolysin.Viruses. 2021 Apr 7;13(4):631. doi: 10.3390/v13040631. Viruses. 2021. PMID: 33917076 Free PMC article.
-
Isolation of a bacteriophage targeting Pseudomonas aeruginosa and exhibits a promising in vivo efficacy.AMB Express. 2023 Jul 26;13(1):79. doi: 10.1186/s13568-023-01582-3. AMB Express. 2023. PMID: 37495819 Free PMC article.
-
Isolation and Characterization of Pseudomonas aeruginosa Phages with a Broad Host Spectrum from Hospital Sewage Systems and Their Therapeutic Effect in a Mouse Model.Am J Trop Med Hyg. 2023 Apr 24;108(6):1220-1226. doi: 10.4269/ajtmh.22-0303. Print 2023 Jun 7. Am J Trop Med Hyg. 2023. PMID: 37094788 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

