Control of Probe Density at DNA Biosensor Surfaces Using Poly(l-lysine) with Appended Reactive Groups
- PMID: 30412384
- PMCID: PMC6302315
- DOI: 10.1021/acs.bioconjchem.8b00733
Control of Probe Density at DNA Biosensor Surfaces Using Poly(l-lysine) with Appended Reactive Groups
Abstract
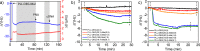
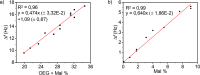
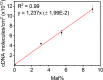
Biosensors and materials for biomedical applications generally require chemical functionalization to bestow their surfaces with desired properties, such as specific molecular recognition and antifouling properties. The use of modified poly(l-lysine) (PLL) polymers with appended oligo(ethylene glycol) (OEG) and thiol-reactive maleimide (Mal) moieties (PLL-OEG-Mal) offers control over the presentation of functional groups. These reactive groups can readily be conjugated to, for example, probes for DNA detection. Here we demonstrate the reliable conjugation of thiol-functionalized peptide nucleic acid (PNA) probes onto predeposited layers of PLL-OEG-Mal and the control over their surface density in the preceding synthetic step of the PLL modification with Mal groups. By monitoring the quartz crystal microbalance (QCM) frequency shifts of the binding of complementary DNA versus the density of Mal moieties grafted to the PLL, a linear relationship between probe density and PLL grafting density was found. Cyclic voltammetry experiments using Methylene Blue-functionalized DNA were performed to establish the absolute probe density values at the biosensor surfaces. These data provided a density of 1.2 × 1012 probes per cm2 per % of grafted Mal, thus confirming the validity of the density control in the synthetic PLL modification step without the need of further surface characterization.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Increasing the Sensitivity of Electrochemical DNA Detection by a Micropillar-Structured Biosensing Surface.Langmuir. 2020 Apr 28;36(16):4272-4279. doi: 10.1021/acs.langmuir.0c00144. Epub 2020 Apr 14. Langmuir. 2020. PMID: 32239946 Free PMC article.
-
Low-fouling, mixed-charge poly-l-lysine polymers with anionic oligopeptide side-chains.J Mater Chem B. 2018 Dec 14;6(46):7662-7673. doi: 10.1039/c8tb01619d. Epub 2018 Nov 5. J Mater Chem B. 2018. PMID: 32254888
-
Influence of pH and Surface Chemistry on Poly(L-lysine) Adsorption onto Solid Supports Investigated by Quartz Crystal Microbalance with Dissipation Monitoring.J Phys Chem B. 2015 Aug 20;119(33):10554-65. doi: 10.1021/acs.jpcb.5b01553. Epub 2015 Aug 7. J Phys Chem B. 2015. PMID: 26061703
-
Effect of peptide secondary structure on adsorption and adsorbed film properties on end-grafted polyethylene oxide layers.Acta Biomater. 2014 Jan;10(1):56-66. doi: 10.1016/j.actbio.2013.09.010. Epub 2013 Sep 21. Acta Biomater. 2014. PMID: 24060880
-
DNA Loading and Release Using Custom-Tailored Poly(l-lysine) Surfaces.ACS Appl Mater Interfaces. 2017 Jul 19;9(28):23370-23378. doi: 10.1021/acsami.7b05024. Epub 2017 Jul 10. ACS Appl Mater Interfaces. 2017. PMID: 28636320
Cited by
-
"Plug-n-Play" Polymer Substrates: Surface Patterning with Reactive-Group-Appended Poly-l-lysine for Biomolecule Adhesion.ACS Appl Polym Mater. 2019 Nov 8;1(11):3165-3173. doi: 10.1021/acsapm.9b00814. Epub 2019 Oct 1. ACS Appl Polym Mater. 2019. PMID: 32954353 Free PMC article.
-
Interface Engineering of "Clickable" Organic Electrochemical Transistors toward Biosensing Devices.ACS Appl Mater Interfaces. 2023 Mar 1;15(8):10885-10896. doi: 10.1021/acsami.2c21493. Epub 2023 Feb 15. ACS Appl Mater Interfaces. 2023. PMID: 36791086 Free PMC article.
-
Enhancement of Probe Density in DNA Sensing by Tuning the Exponential Growth Regime of Polyelectrolyte Multilayers.Chem Mater. 2020 Nov 10;32(21):9155-9166. doi: 10.1021/acs.chemmater.0c02454. Epub 2020 Oct 26. Chem Mater. 2020. PMID: 33191977 Free PMC article.
-
Covalent organic frameworks in cancer theranostics: advancing biomarker detection and tumor-targeted therapy.Arch Pharm Res. 2025 Mar;48(3):183-211. doi: 10.1007/s12272-025-01536-2. Epub 2025 Mar 22. Arch Pharm Res. 2025. PMID: 40119211 Review.
-
Identification of novel human IgE-binding peptides from a phage display library for total IgE detection.Sci Rep. 2025 Jul 31;15(1):27986. doi: 10.1038/s41598-025-12574-7. Sci Rep. 2025. PMID: 40745439 Free PMC article.
References
-
- Seeman C. N.; Sleiman F. H. (2017) DNA nanotechnology. Nat. Rev. Mater. 3, 17068.10.1038/natrevmats.2017.68. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

