Comparative mRNA and miRNA transcriptome analysis of a mouse model of IGFIR-driven lung cancer
- PMID: 30412601
- PMCID: PMC6226179
- DOI: 10.1371/journal.pone.0206948
Comparative mRNA and miRNA transcriptome analysis of a mouse model of IGFIR-driven lung cancer
Abstract
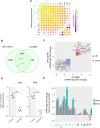
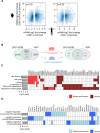
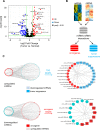
Mouse models of cancer play an important role in elucidating the molecular mechanisms that contribute to tumorigenesis. The extent to which these models resemble one another and their human counterparts at the molecular level is critical in understanding tumorigenesis. In this study, we carried out a comparative gene expression analysis to generate a detailed molecular portrait of a transgenic mouse model of IGFIR-driven lung cancer. IGFIR-driven tumors displayed a strong resemblance with established mouse models of lung adenocarcinoma, particularly EGFR-driven models highlighted by elevated levels of the EGFR ligands Ereg and Areg. Cross-species analysis revealed a shared increase in human lung adenocarcinoma markers including Nkx2.1 and Napsa as well as alterations in a subset of genes with oncogenic and tumor suppressive properties such as Aurka, Ret, Klf4 and Lats2. Integrated miRNA and mRNA analysis in IGFIR-driven tumors identified interaction pairs with roles in ErbB signaling while cross-species analysis revealed coordinated expression of a subset of conserved miRNAs and their targets including miR-21-5p (Reck, Timp3 and Tgfbr3). Overall, these findings support the use of SPC-IGFIR mice as a model of human lung adenocarcinoma and provide a comprehensive knowledge base to dissect the molecular pathogenesis of tumor initiation and progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. Elsevier; 2015;10: 1243–1260. 10.1097/JTO.0000000000000630 - DOI - PubMed
-
- Vikis HG, Rymaszewski AL, Tichelaar JW. Mouse models of chemically-induced lung carcinogenesis. Front Biosci (Elite Ed). 2013;5: 939–46. Available: http://www.ncbi.nlm.nih.gov/pubmed/23747909 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

