Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer's disease
- PMID: 30413934
- PMCID: PMC6358498
- DOI: 10.1007/s00401-018-1928-6
Dissecting the genetic relationship between cardiovascular risk factors and Alzheimer's disease
Abstract
Cardiovascular (CV)- and lifestyle-associated risk factors (RFs) are increasingly recognized as important for Alzheimer's disease (AD) pathogenesis. Beyond the ε4 allele of apolipoprotein E (APOE), comparatively little is known about whether CV-associated genes also increase risk for AD. Using large genome-wide association studies and validated tools to quantify genetic overlap, we systematically identified single nucleotide polymorphisms (SNPs) jointly associated with AD and one or more CV-associated RFs, namely body mass index (BMI), type 2 diabetes (T2D), coronary artery disease (CAD), waist hip ratio (WHR), total cholesterol (TC), triglycerides (TG), low-density (LDL) and high-density lipoprotein (HDL). In fold enrichment plots, we observed robust genetic enrichment in AD as a function of plasma lipids (TG, TC, LDL, and HDL); we found minimal AD genetic enrichment conditional on BMI, T2D, CAD, and WHR. Beyond APOE, at conjunction FDR < 0.05 we identified 90 SNPs on 19 different chromosomes that were jointly associated with AD and CV-associated outcomes. In meta-analyses across three independent cohorts, we found four novel loci within MBLAC1 (chromosome 7, meta-p = 1.44 × 10-9), MINK1 (chromosome 17, meta-p = 1.98 × 10-7) and two chromosome 11 SNPs within the MTCH2/SPI1 region (closest gene = DDB2, meta-p = 7.01 × 10-7 and closest gene = MYBPC3, meta-p = 5.62 × 10-8). In a large 'AD-by-proxy' cohort from the UK Biobank, we replicated three of the four novel AD/CV pleiotropic SNPs, namely variants within MINK1, MBLAC1, and DDB2. Expression of MBLAC1, SPI1, MINK1 and DDB2 was differentially altered within postmortem AD brains. Beyond APOE, we show that the polygenic component of AD is enriched for lipid-associated RFs. We pinpoint a subset of cardiovascular-associated genes that strongly increase the risk for AD. Our collective findings support a disease model in which cardiovascular biology is integral to the development of clinical AD in a subset of individuals.
Keywords: Alzheimer’s disease; Cardiovascular; Genetic pleiotropy; Lipids; Polygenic enrichment.
Conflict of interest statement
Figures
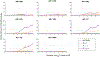

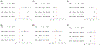


References
Publication types
MeSH terms
Substances
Grants and funding
- 223273/Norges Forskningsråd/International
- U24 AG021886/AG/NIA NIH HHS/United States
- 237250/EU JPND/Norges Forskningsråd/International
- RF1 AG054023/AG/NIA NIH HHS/United States
- K01 AG046374/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- 225989/Norges Forskningsråd/International
- RF1 AG015473/AG/NIA NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 213837/Norges Forskningsråd/International
- R01 AG015819/AG/NIA NIH HHS/United States
- K24 AG035007/AG/NIA NIH HHS/United States
- K01 AG049152/AG/NIA NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- U01 AG032984/AG/NIA NIH HHS/United States
- U24 AG056270/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

