Detection of ribonucleoside modifications by liquid chromatography coupled with mass spectrometry
- PMID: 30414470
- PMCID: PMC6401287
- DOI: 10.1016/j.bbagrm.2018.10.012
Detection of ribonucleoside modifications by liquid chromatography coupled with mass spectrometry
Abstract
A small set of ribonucleoside modifications have been found in different regions of mRNA including the open reading frame. Accurate detection of these specific modifications is critical to understanding their modulatory roles in facilitating mRNA maturation, translation and degradation. While transcriptome-wide next-generation sequencing (NGS) techniques could provide exhaustive information about the sites of one specific or class of modifications at a time, recent investigations strongly indicate cautionary interpretation due to the appearance of false positives. Therefore, it is suggested that NGS-based modification data can only be treated as predicted sites and their existence need to be validated by orthogonal methods. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is an analytical technique that can yield accurate and reproducible information about the qualitative and quantitative characteristics of ribonucleoside modifications. Here, we review the recent advancements in LC-MS/MS technology that could help in securing accurate, gold-standard quality information about the resident post-transcriptional modifications of mRNA.
Keywords: LC-MS; Nucleoside analysis; Positional isomers; Post-transcriptional modifications; RNA modification mapping.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest: The authors declare no conflict of interest with this manuscript.
Figures
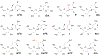


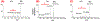
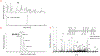
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

