Requirement of Pitx2 for skeletal muscle homeostasis
- PMID: 30414844
- PMCID: PMC6289786
- DOI: 10.1016/j.ydbio.2018.11.001
Requirement of Pitx2 for skeletal muscle homeostasis
Abstract
Skeletal muscle is generated by the successive incorporation of primary (embryonic), secondary (fetal), and tertiary (adult) fibers into muscle. Conditional excision of Pitx2 function by an MCKCre driver resulted in animals with histological and ultrastructural defects in P30 muscles and fibers, respectively. Mutant muscle showed severe reduction in mitochondria and FoxO3-mediated mitophagy. Both oxidative and glycolytic energy metabolism were reduced. Conditional excision was limited to fetal muscle fibers after the G1-G0 transition and resulted in altered MHC, Rac1, MEF2a, and alpha-tubulin expression within these fibers. The onset of excision, monitored by a nuclear reporter gene, was observed as early as E16. Muscle at this stage was already severely malformed, but appeared to recover by P30 by the expansion of adjoining larger fibers. Our studies demonstrate that the homeodomain transcription factor Pitx2 has a postmitotic role in maintaining skeletal muscle integrity and energy homeostasis in fetal muscle fibers.
Keywords: Autophagy; Mitochondria; Mouse; Pitx2; Skeletal muscle.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing interests
No competing interests declared.
Figures

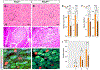
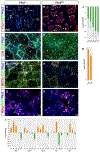

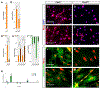
References
-
- Abu-Elmagd M, Robson L, Sweetman D, Hadley J, Francis-West P, Münsterberg A, 2010. Wnt/Lef1 signaling acts via Pitx2 to regulate somite myogenesis. Dev. Biol 337, 211–219. doi:10.1016/j.ydbio.2009.10.023 - DOI - PubMed
-
- Ai D, Liu W, Ma L, Dong F, Lu M-F, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, Black BL, Brown NA, Martin JF, 2006. Pitx2 regulates cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Developmental Biology 296, 437–449. doi:10.1016/j.ydbio.2006.06.009 - DOI - PMC - PubMed
-
- Alvares LE, Schubert FR, Thorpe C, Mootoosamy RC, Cheng L, Parkyn G, Lumsden A, Dietrich S, 2003. Intrinsic, Hox-dependent cues determine the fate of skeletal muscle precursors. DEVCEL 5, 379–390. - PubMed
-
- Bajanca F, Luz M, Raymond K, Martins GG, Sonnenberg A, Tajbakhsh S, Buckingham M, Thorsteinsdóttir S, 2006. Integrin alpha6beta1-laminin interactions regulate early myotome formation in the mouse embryo. Development 133, 1635–1644. doi:10.1242/dev.02336 - DOI - PubMed
-
- Berkes CA, Tapscott SJ, 2005. MyoD and the transcriptional control of myogenesis. Seminars in Cell and Developmental Biology 16, 585–595. doi:10.1016/j.semcdb.2005.07.006 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

