Gain of CTCF-Anchored Chromatin Loops Marks the Exit from Naive Pluripotency
- PMID: 30414923
- PMCID: PMC6327227
- DOI: 10.1016/j.cels.2018.09.003
Gain of CTCF-Anchored Chromatin Loops Marks the Exit from Naive Pluripotency
Abstract
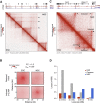
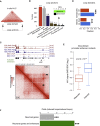
The genome of pluripotent stem cells adopts a unique three-dimensional architecture featuring weakly condensed heterochromatin and large nucleosome-free regions. Yet, it is unknown whether structural loops and contact domains display characteristics that distinguish embryonic stem cells (ESCs) from differentiated cell types. We used genome-wide chromosome conformation capture and super-resolution imaging to determine nuclear organization in mouse ESC and neural stem cell (NSC) derivatives. We found that loss of pluripotency is accompanied by widespread gain of structural loops. This general architectural change correlates with enhanced binding of CTCF and cohesins and more pronounced insulation of contacts across chromatin boundaries in lineage-committed cells. Reprogramming NSCs to pluripotency restores the unique features of ESC domain topology. Domains defined by the anchors of loops established upon differentiation are enriched for developmental genes. Chromatin loop formation is a pervasive structural alteration to the genome that accompanies exit from pluripotency and delineates the spatial segregation of developmentally regulated genes.
Keywords: CTCF; CTCF loops; chromatin architecture; chromatin loops; chromatin structure; differentiation; pluripotency; topologically associating domains.
Published by Elsevier Inc.
Figures





Similar articles
-
SRF promotes long-range chromatin loop formation and stem cell pluripotency.Cell Rep. 2024 Oct 22;43(10):114846. doi: 10.1016/j.celrep.2024.114846. Epub 2024 Oct 10. Cell Rep. 2024. PMID: 39392751
-
Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver.Elife. 2018 May 14;7:e34077. doi: 10.7554/eLife.34077. Elife. 2018. PMID: 29757144 Free PMC article.
-
Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus.Proc Natl Acad Sci U S A. 2011 May 3;108(18):7391-6. doi: 10.1073/pnas.1018279108. Epub 2011 Apr 18. Proc Natl Acad Sci U S A. 2011. PMID: 21502535 Free PMC article.
-
Function and regulation of chromatin insulators in dynamic genome organization.Curr Opin Cell Biol. 2019 Jun;58:61-68. doi: 10.1016/j.ceb.2019.02.001. Epub 2019 Mar 12. Curr Opin Cell Biol. 2019. PMID: 30875678 Free PMC article. Review.
-
Chromatin Architecture in the Fly: Living without CTCF/Cohesin Loop Extrusion?: Alternating Chromatin States Provide a Basis for Domain Architecture in Drosophila.Bioessays. 2019 Sep;41(9):e1900048. doi: 10.1002/bies.201900048. Epub 2019 Jul 1. Bioessays. 2019. PMID: 31264253 Review.
Cited by
-
Genetic control of the pluripotency epigenome determines differentiation bias in mouse embryonic stem cells.EMBO J. 2022 Dec 17;41(2):e109445. doi: 10.15252/embj.2021109445. Epub 2021 Dec 21. EMBO J. 2022. PMID: 34931323 Free PMC article.
-
CTCF deletion alters the pluripotency and DNA methylation profile of human iPSCs.Front Cell Dev Biol. 2023 Nov 30;11:1302448. doi: 10.3389/fcell.2023.1302448. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38099298 Free PMC article.
-
CTCF confers local nucleosome resiliency after DNA replication and during mitosis.Elife. 2019 Oct 10;8:e47898. doi: 10.7554/eLife.47898. Elife. 2019. PMID: 31599722 Free PMC article.
-
Chromatin insulation dynamics in glioblastoma: challenges and future perspectives of precision oncology.Clin Epigenetics. 2021 Jul 31;13(1):150. doi: 10.1186/s13148-021-01139-w. Clin Epigenetics. 2021. PMID: 34332627 Free PMC article.
-
Epigenetic regulation in adult neural stem cells.Front Cell Dev Biol. 2024 Jan 31;12:1331074. doi: 10.3389/fcell.2024.1331074. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38357000 Free PMC article. Review.
References
-
- Banterle N., Bui K.H., Lemke E.A., Beck M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol. 2013;183:363–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

