Reconstructing the Deep Population History of Central and South America
- PMID: 30415837
- PMCID: PMC6327247
- DOI: 10.1016/j.cell.2018.10.027
Reconstructing the Deep Population History of Central and South America
Abstract
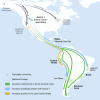
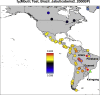
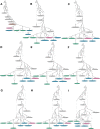
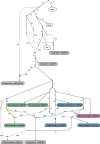
We report genome-wide ancient DNA from 49 individuals forming four parallel time transects in Belize, Brazil, the Central Andes, and the Southern Cone, each dating to at least ∼9,000 years ago. The common ancestral population radiated rapidly from just one of the two early branches that contributed to Native Americans today. We document two previously unappreciated streams of gene flow between North and South America. One affected the Central Andes by ∼4,200 years ago, while the other explains an affinity between the oldest North American genome associated with the Clovis culture and the oldest Central and South Americans from Chile, Brazil, and Belize. However, this was not the primary source for later South Americans, as the other ancient individuals derive from lineages without specific affinity to the Clovis-associated genome, suggesting a population replacement that began at least 9,000 years ago and was followed by substantial population continuity in multiple regions.
Keywords: Central America; South America; anthropology; archaeology; population genetics.
Published by Elsevier Inc.
Figures








Comment in
-
Native American Genomic Diversity through Ancient DNA.Cell. 2018 Nov 15;175(5):1173-1174. doi: 10.1016/j.cell.2018.10.058. Cell. 2018. PMID: 30445034
References
-
- Cardich A. Volume 3. Centro Argentino de Estudios Prehistóricos; 1964. (Lauricocha: Fundamentos para una prehistoria de los Andes centrales).
-
- Chala-Aldana D., Bocherens H., Miller C., Moore K., Hodgins G., Rademaker K. Investigating mobility and highland occupation strategies during the Early Holocene at the Cuncaicha rock shelter through strontium and oxygen isotopes. J. Archaeol. Sci. Rep. 2018;19:811–827.

