Characterization of the Microbiome at the World's Largest Potable Water Reuse Facility
- PMID: 30416489
- PMCID: PMC6212505
- DOI: 10.3389/fmicb.2018.02435
Characterization of the Microbiome at the World's Largest Potable Water Reuse Facility
Abstract
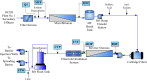
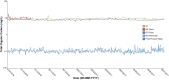
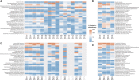
Conventional water resources are not sufficient in many regions to meet the needs of growing populations. Due to cyclical weather cycles, drought, and climate change, water stress has increased worldwide including in Southern California, which serves as a model for regions that integrate reuse of wastewater for both potable and non-potable use. The Orange County Water District (OCWD) Advanced Water Purification Facility (AWPF) is a highly engineered system designed to treat and produce up to 100 million gallons per day (MGD) of purified water from a municipal wastewater source for potable reuse. Routine facility microbial water quality analysis is limited to standard indicators at this and similar facilities. Given recent advances in high throughput DNA sequencing techniques, complete microbial profiling of communities in water samples is now possible. By using 16S/18S rRNA gene sequencing, metagenomic and metatranscriptomic sequencing coupled to a highly accurate identification method along with 16S rRNA gene qPCR, we describe a detailed view of the total microbial community throughout the facility. The total bacterial load of the water at stages of the treatment train ranged from 3.02 × 106 copies in source, unchlorinated wastewater feed to 5.49 × 101 copies of 16S rRNA gene/mL after treatment (consisting of microfiltration, reverse osmosis, and ultraviolet/advanced oxidation). Microbial diversity and load decreased by several orders of magnitude after microfiltration and reverse osmosis treatment, falling to almost non-detectable levels that more closely resembled controls of molecular grade laboratory water than the biomass detected in the source water. The presence of antibiotic resistance genes and viruses was also greatly reduced. Overall, system design performance was achieved, and comprehensive microbial community analysis was found to enable a more complete characterization of the water/wastewater microbial signature.
Keywords: metagenomics; metatranscriptomics; pathogens; water purification; water reuse.
Figures



References
-
- Aieta E. M., Berg J. D. (1986). A review of chlorine dioxide in drinking-water treatment. J. Am. Water Works Assoc. 78 62–72. 10.1002/j.1551-8833.1986.tb05766.x - DOI
-
- Amha Y. M., Anwar M. Z., Kumaraswamy R., Henschel A., Ahmad F. (2017). Mycobacteria in municipal wastewater treatment and reuse: microbial diversity for screening the occurrence of clinically and environmentally relevant species in arid regions. Environ. Sci. Technol. 51 3048–3056. 10.1021/acs.est.6b05580 - DOI - PubMed
LinkOut - more resources
Full Text Sources

