Metabolite sensing and signaling in cell metabolism
- PMID: 30416760
- PMCID: PMC6224561
- DOI: 10.1038/s41392-018-0024-7
Metabolite sensing and signaling in cell metabolism
Abstract
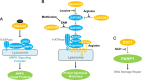
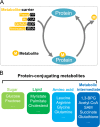
Metabolite sensing is one of the most fundamental biological processes. During evolution, multilayered mechanisms developed to sense fluctuations in a wide spectrum of metabolites, including nutrients, to coordinate cellular metabolism and biological networks. To date, AMPK and mTOR signaling are among the best-understood metabolite-sensing and signaling pathways. Here, we propose a sensor-transducer-effector model to describe known mechanisms of metabolite sensing and signaling. We define a metabolite sensor by its specificity, dynamicity, and functionality. We group the actions of metabolite sensing into three different modes: metabolite sensor-mediated signaling, metabolite-sensing module, and sensing by conjugating. With these modes of action, we provide a systematic view of how cells sense sugars, lipids, amino acids, and metabolic intermediates. In the future perspective, we suggest a systematic screen of metabolite-sensing macromolecules, high-throughput discovery of biomacromolecule-metabolite interactomes, and functional metabolomics to advance our knowledge of metabolite sensing and signaling. Most importantly, targeting metabolite sensing holds great promise in therapeutic intervention of metabolic diseases and in improving healthy aging.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

