Multi-faceted computational assessment of risk and progression in oligodendroglioma implicates NOTCH and PI3K pathways
- PMID: 30417117
- PMCID: PMC6219505
- DOI: 10.1038/s41698-018-0067-9
Multi-faceted computational assessment of risk and progression in oligodendroglioma implicates NOTCH and PI3K pathways
Abstract
Oligodendrogliomas are diffusely infiltrative gliomas defined by IDH-mutation and co-deletion of 1p/19q. They have highly variable clinical courses, with survivals ranging from 6 months to over 20 years, but little is known regarding the pathways involved with their progression or optimal markers for stratifying risk. We utilized machine-learning approaches with genomic data from The Cancer Genome Atlas to objectively identify molecular factors associated with clinical outcomes of oligodendroglioma and extended these findings to study signaling pathways implicated in oncogenesis and clinical endpoints associated with glioma progression. Our multi-faceted computational approach uncovered key genetic alterations associated with disease progression and shorter survival in oligodendroglioma and specifically identified Notch pathway inactivation and PI3K pathway activation as the most strongly associated with MRI and pathology findings of advanced disease and poor clinical outcome. Our findings that Notch pathway inactivation and PI3K pathway activation are associated with advanced disease and survival risk will pave the way for clinically relevant markers of disease progression and therapeutic targets to improve clinical outcomes. Furthermore, our approach demonstrates the strength of machine learning and computational methods for identifying genetic events critical to disease progression in the era of big data and precision medicine.
Conflict of interest statement
The authors declare no competing interests.
Figures
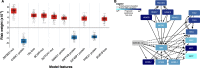
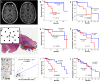

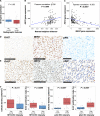
References
-
- Louis, DN, Ohgaki, H., Wiestler, OD, Cavenee, WK. World Health Organization Histological Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer, France (2016).
Grants and funding
LinkOut - more resources
Full Text Sources

