The sacrificial inactivation of the blue-light photosensor cryptochrome from Drosophila melanogaster
- PMID: 30417904
- PMCID: PMC6250122
- DOI: 10.1039/c8cp04671a
The sacrificial inactivation of the blue-light photosensor cryptochrome from Drosophila melanogaster
Abstract
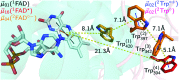
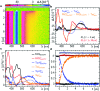
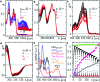
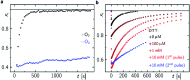
Drosophila melanogaster cryptochrome functions as the primary blue-light receptor that mediates circadian photo entrainment. Absorption of a photon leads to reduction of the protein-bound FAD via consecutive electron transfer along a conserved tryptophan tetrad resembling the signalling state required for conformational changes and induction of subsequent signalling cascades. However, how the initial photochemistry and subsequent dark processes leading to downstream signalling are linked to each other at the molecular level is still poorly understood. Here, we investigated in detail the initial photochemical events in DmCRY by time-resolved and stationary absorption spectroscopy combined with quantum chemical and molecular dynamics calculations. We resolved the early events along the conserved tryptophan tetrad and the final deprotonation of the terminal tryptophanyl radical cation. These initial events lead to conformational changes, such as the known C-terminal tail release, Trp decomposition, and finally FAD release providing evidence that DmCRY does not undergo a photocycle. We propose that light is a negative regulator of DmCRY stability even under in vitro conditions where the proteasomal machinery is missing, that is in line with its biological function, i.e. entrainment of the circadian clock.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

